

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Programmed cell death 1 ligand 1 [19-238]/protein 1 [25-167] | ||
| Ligand | BDBM452136 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | Biochemical Protein-Protein Interaction Assay | ||
| IC50 | 0.064±n/a nM | ||
| Citation |  Aktoudianakis, E; Cho, A; Du, Z; Graupe, M; Lad, LT; Machicao Tello, PA; Medley, JW; Metobo, SE; Mukherjee, PK; Naduthambi, D; Parkhill, EQ; Phillips, BW; Simonovich, SP; Squires, NH; Wang, P; Watkins, WJ; Xu, J; Yang, KS; Ziebenhaus, CA PD-1/PD-L1 inhibitors US Patent US10710986 Publication Date 7/14/2020 Aktoudianakis, E; Cho, A; Du, Z; Graupe, M; Lad, LT; Machicao Tello, PA; Medley, JW; Metobo, SE; Mukherjee, PK; Naduthambi, D; Parkhill, EQ; Phillips, BW; Simonovich, SP; Squires, NH; Wang, P; Watkins, WJ; Xu, J; Yang, KS; Ziebenhaus, CA PD-1/PD-L1 inhibitors US Patent US10710986 Publication Date 7/14/2020 | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Programmed cell death 1 ligand 1 [19-238]/protein 1 [25-167] | |||
| Name: | Programmed cell death 1 ligand 1 [19-238]/protein 1 [25-167] | ||
| Synonyms: | PD-1/PD-L1 | ||
| Type: | Protein | ||
| Mol. Mass.: | n/a | ||
| Description: | n/a | ||
| Components: | This complex has 2 components. | ||
| Component 1 | |||
| Name: | Programmed cell death protein 1 [25-167] | ||
| Synonyms: | PD1 | PDCD1 | PDCD1_HUMAN | Programmed cell death protein 1 (25-167) | Programmed cell death protein 1 (aa25-167) | Protein PD-1 | hPD-1 | ||
| Type: | Enzyme Catalytic Domain | ||
| Mol. Mass.: | 15954.64 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Q15116 (aa25-167) | ||
| Residue: | 143 | ||
| Sequence: |
| ||
| Component 2 | |||
| Name: | Programmed cell death 1 ligand 1 [19-238] | ||
| Synonyms: | B7H1 | CD274 | PD-L1 | PD1L1_HUMAN | PDCD1 ligand 1 | PDCD1L1 | PDCD1LG1 | PDL1 | Programmed cell death 1 ligand 1 (aa19-238) | Programmed death ligand 1 | hPD-L1 | ||
| Type: | Enzyme Catalytic Domain | ||
| Mol. Mass.: | 25191.31 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Q9NZQ7 (aa19-238) | ||
| Residue: | 220 | ||
| Sequence: |
| ||
| BDBM452136 | |||
| n/a | |||
| Name | BDBM452136 | ||
| Synonyms: | US10710986, Example 275 | US11555029, No. 275 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C34H37Cl2N7O4 | ||
| Mol. Mass. | 678.608 | ||
| SMILES | COc1nc(cnc1CNC[C@H]1C[C@H](O)C1)-c1cccc(c1Cl)-c1cccc(c1Cl)-c1cnc(CNC[C@@H]2CCC(=O)N2)c(OC)n1 |r,wU:37.40,11.11,wD:13.14,(8.86,-1.93,;8.86,-.39,;7.52,.38,;6.19,-.39,;4.86,.38,;4.86,1.92,;6.19,2.69,;7.52,1.92,;8.86,2.69,;10.19,1.92,;11.53,2.7,;12.86,1.93,;14.19,2.7,;14.96,1.36,;16.45,.96,;13.63,.59,;3.52,-.39,;3.52,-1.93,;2.19,-2.7,;.86,-1.93,;.86,-.39,;2.19,.38,;2.19,1.92,;-.48,.38,;-.48,1.92,;-1.81,2.69,;-3.15,1.92,;-3.15,.38,;-1.81,-.39,;-1.81,-1.93,;-4.48,-.39,;-4.48,-1.93,;-5.81,-2.7,;-7.15,-1.93,;-8.48,-2.7,;-9.81,-1.93,;-11.15,-2.7,;-12.48,-1.93,;-12.64,-.39,;-14.15,-.07,;-14.92,-1.41,;-16.45,-1.57,;-13.89,-2.55,;-7.15,-.39,;-8.48,.38,;-8.48,1.92,;-5.81,.38,)| | ||
| Structure | 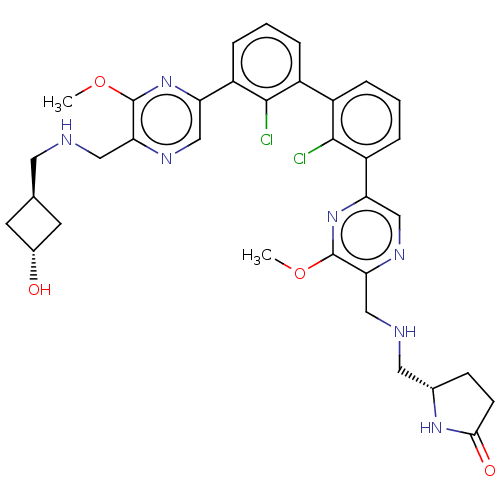
| ||