| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Serine protease 1 |
|---|
| Ligand | BDBM14152 |
|---|
| Substrate/Competitor | BDBM12679 |
|---|
| Meas. Tech. | Enzyme Assay and Determination of the Inhibition Constants |
|---|
| Ki | 230±n/a nM |
|---|
| Citation |  Katz, BA; Sprengeler, PA; Luong, C; Verner, E; Elrod, K; Kirtley, M; Janc, J; Spencer, JR; Breitenbucher, JG; Hui, H; McGee, D; Allen, D; Martelli, A; Mackman, RL Engineering inhibitors highly selective for the S1 sites of Ser190 trypsin-like serine protease drug targets. Chem Biol8:1107-21 (2001) [PubMed] Article Katz, BA; Sprengeler, PA; Luong, C; Verner, E; Elrod, K; Kirtley, M; Janc, J; Spencer, JR; Breitenbucher, JG; Hui, H; McGee, D; Allen, D; Martelli, A; Mackman, RL Engineering inhibitors highly selective for the S1 sites of Ser190 trypsin-like serine protease drug targets. Chem Biol8:1107-21 (2001) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Serine protease 1 |
|---|
| Name: | Serine protease 1 |
|---|
| Synonyms: | Beta-Trypsin | Cationic trypsin | PRSS1 | TRP1 | TRY1 | TRY1_BOVIN | TRYP1 | Trypsin | Trypsin I |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 25790.52 |
|---|
| Organism: | Bos taurus (bovine) |
|---|
| Description: | P00760 |
|---|
| Residue: | 246 |
|---|
| Sequence: | MKTFIFLALLGAAVAFPVDDDDKIVGGYTCGANTVPYQVSLNSGYHFCGGSLINSQWVVS
AAHCYKSGIQVRLGEDNINVVEGNEQFISASKSIVHPSYNSNTLNNDIMLIKLKSAASLN
SRVASISLPTSCASAGTQCLISGWGNTKSSGTSYPDVLKCLKAPILSDSSCKSAYPGQIT
SNMFCAGYLEGGKDSCQGDSGGPVVCSGKLQGIVSWGSGCAQKNKPGVYTKVCNYVSWIK
QTIASN
|
|
|
|---|
| BDBM14152 |
|---|
| BDBM12679 |
|---|
| Name | BDBM14152 |
|---|
| Synonyms: | 6-CHLORO-2-(2-HYDROXY-BIPHENYL-3-YL)-1H-INDOLE-5-CARBOXAMIDINE | APC-10302 | CA-14 | CRA-10302 | {amino[6-chloro-2-(2-hydroxy-3-phenylphenyl)-1H-indol-5-yl]methylidene}azanium |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C21H17ClN3O |
|---|
| Mol. Mass. | 362.832 |
|---|
| SMILES | NC(=[NH2+])c1cc2cc([nH]c2cc1Cl)-c1cccc(-c2ccccc2)c1O |
|---|
| Structure | 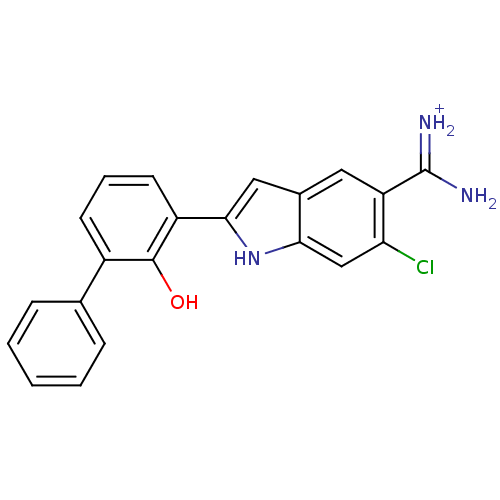
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Katz, BA; Sprengeler, PA; Luong, C; Verner, E; Elrod, K; Kirtley, M; Janc, J; Spencer, JR; Breitenbucher, JG; Hui, H; McGee, D; Allen, D; Martelli, A; Mackman, RL Engineering inhibitors highly selective for the S1 sites of Ser190 trypsin-like serine protease drug targets. Chem Biol8:1107-21 (2001) [PubMed] Article
Katz, BA; Sprengeler, PA; Luong, C; Verner, E; Elrod, K; Kirtley, M; Janc, J; Spencer, JR; Breitenbucher, JG; Hui, H; McGee, D; Allen, D; Martelli, A; Mackman, RL Engineering inhibitors highly selective for the S1 sites of Ser190 trypsin-like serine protease drug targets. Chem Biol8:1107-21 (2001) [PubMed] Article