| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Peroxisome proliferator-activated receptor alpha |
|---|
| Ligand | BDBM24566 |
|---|
| Substrate/Competitor | BDBM10852 |
|---|
| Meas. Tech. | Cell-Based Transcription Assay |
|---|
| EC50 | 1620±340 nM |
|---|
| Comments | 100 % activation. |
|---|
| Citation |  Montanari, R; Saccoccia, F; Scotti, E; Crestani, M; Godio, C; Gilardi, F; Loiodice, F; Fracchiolla, G; Laghezza, A; Tortorella, P; Lavecchia, A; Novellino, E; Mazza, F; Aschi, M; Pochetti, G Crystal structure of the peroxisome proliferator-activated receptor gamma (PPARgamma) ligand binding domain complexed with a novel partial agonist: a new region of the hydrophobic pocket could be exploited for drug design. J Med Chem51:7768-76 (2008) [PubMed] Article Montanari, R; Saccoccia, F; Scotti, E; Crestani, M; Godio, C; Gilardi, F; Loiodice, F; Fracchiolla, G; Laghezza, A; Tortorella, P; Lavecchia, A; Novellino, E; Mazza, F; Aschi, M; Pochetti, G Crystal structure of the peroxisome proliferator-activated receptor gamma (PPARgamma) ligand binding domain complexed with a novel partial agonist: a new region of the hydrophobic pocket could be exploited for drug design. J Med Chem51:7768-76 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Peroxisome proliferator-activated receptor alpha |
|---|
| Name: | Peroxisome proliferator-activated receptor alpha |
|---|
| Synonyms: | NR1C1 | Nuclear receptor subfamily 1 group C member 1 | PPAR | PPAR alpha/gamma | PPAR-alpha | PPARA | PPARA_HUMAN | Peroxisome Proliferator-Activated Receptor alpha | Peroxisome proliferator-activated receptor | Peroxisome proliferator-activated receptor alpha (PPAR alpha) |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 52222.08 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q07869 |
|---|
| Residue: | 468 |
|---|
| Sequence: | MVDTESPLCPLSPLEAGDLESPLSEEFLQEMGNIQEISQSIGEDSSGSFGFTEYQYLGSC
PGSDGSVITDTLSPASSPSSVTYPVVPGSVDESPSGALNIECRICGDKASGYHYGVHACE
GCKGFFRRTIRLKLVYDKCDRSCKIQKKNRNKCQYCRFHKCLSVGMSHNAIRFGRMPRSE
KAKLKAEILTCEHDIEDSETADLKSLAKRIYEAYLKNFNMNKVKARVILSGKASNNPPFV
IHDMETLCMAEKTLVAKLVANGIQNKEAEVRIFHCCQCTSVETVTELTEFAKAIPGFANL
DLNDQVTLLKYGVYEAIFAMLSSVMNKDGMLVAYGNGFITREFLKSLRKPFCDIMEPKFD
FAMKFNALELDDSDISLFVAAIICCGDRPGLLNVGHIEKMQEGIVHVLRLHLQSNHPDDI
FLFPKLLQKMADLRQLVTEHAQLVQIIKKTESDAALHPLLQEIYRDMY
|
|
|
|---|
| BDBM24566 |
|---|
| BDBM10852 |
|---|
| Name | BDBM24566 |
|---|
| Synonyms: | 2-({4-chloro-6-[(2,3-dimethylphenyl)amino]pyrimidin-2-yl}sulfanyl)acetic acid | CHEMBL295416 | JMC515449 Compound 1 | Pirinixic acid | Wyeth 14,643 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C14H14ClN3O2S |
|---|
| Mol. Mass. | 323.798 |
|---|
| SMILES | Cc1cccc(Nc2cc(Cl)nc(SCC(O)=O)n2)c1C |
|---|
| Structure | 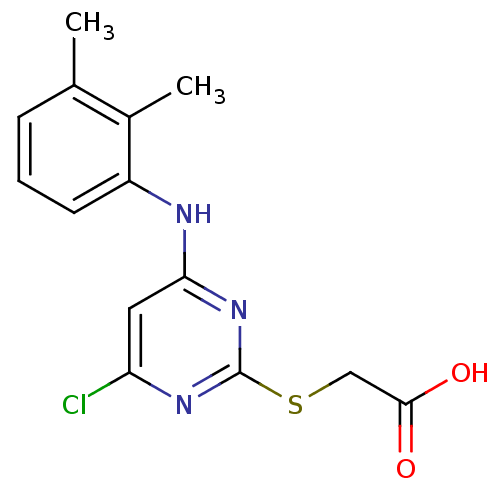
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Montanari, R; Saccoccia, F; Scotti, E; Crestani, M; Godio, C; Gilardi, F; Loiodice, F; Fracchiolla, G; Laghezza, A; Tortorella, P; Lavecchia, A; Novellino, E; Mazza, F; Aschi, M; Pochetti, G Crystal structure of the peroxisome proliferator-activated receptor gamma (PPARgamma) ligand binding domain complexed with a novel partial agonist: a new region of the hydrophobic pocket could be exploited for drug design. J Med Chem51:7768-76 (2008) [PubMed] Article
Montanari, R; Saccoccia, F; Scotti, E; Crestani, M; Godio, C; Gilardi, F; Loiodice, F; Fracchiolla, G; Laghezza, A; Tortorella, P; Lavecchia, A; Novellino, E; Mazza, F; Aschi, M; Pochetti, G Crystal structure of the peroxisome proliferator-activated receptor gamma (PPARgamma) ligand binding domain complexed with a novel partial agonist: a new region of the hydrophobic pocket could be exploited for drug design. J Med Chem51:7768-76 (2008) [PubMed] Article