| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Peroxisome proliferator-activated receptor alpha |
|---|
| Ligand | BDBM28800 |
|---|
| Substrate/Competitor | BDBM10852 |
|---|
| Meas. Tech. | Cell-Based Transcription Assay |
|---|
| EC50 | 317±34 nM |
|---|
| Comments | 84 +/- 10 efficacy. |
|---|
| Citation |  Mukherjee, R; Locke, KT; Miao, B; Meyers, D; Monshizadegan, H; Zhang, R; Search, D; Grimm, D; Flynn, M; O'Malley, KM; Zhang, L; Li, J; Shi, Y; Kennedy, LJ; Blanar, M; Cheng, PT; Tino, J; Srivastava, RA Novel peroxisome proliferator-activated receptor alpha agonists lower low-density lipoprotein and triglycerides, raise high-density lipoprotein, and synergistically increase cholesterol excretion with a liver X receptor agonist. J Pharmacol Exp Ther327:716-26 (2008) [PubMed] Article Mukherjee, R; Locke, KT; Miao, B; Meyers, D; Monshizadegan, H; Zhang, R; Search, D; Grimm, D; Flynn, M; O'Malley, KM; Zhang, L; Li, J; Shi, Y; Kennedy, LJ; Blanar, M; Cheng, PT; Tino, J; Srivastava, RA Novel peroxisome proliferator-activated receptor alpha agonists lower low-density lipoprotein and triglycerides, raise high-density lipoprotein, and synergistically increase cholesterol excretion with a liver X receptor agonist. J Pharmacol Exp Ther327:716-26 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Peroxisome proliferator-activated receptor alpha |
|---|
| Name: | Peroxisome proliferator-activated receptor alpha |
|---|
| Synonyms: | Nr1c1 | Nuclear receptor subfamily 1 group C member 1 | PPAR-alpha | PPARA_RAT | Peroxisome Proliferator-Activated Receptor alpha | Ppar | Ppara |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 52374.28 |
|---|
| Organism: | Rattus norvegicus |
|---|
| Description: | ChEMBL_834194 |
|---|
| Residue: | 468 |
|---|
| Sequence: | MVDTESPICPLSPLEADDLESPLSEEFLQEMGNIQEISQSLGEESSGSFSFADYQYLGSC
PGSEGSVITDTLSPASSPSSVSCPAVPTSTDESPGNALNIECRICGDKASGYHYGVHACE
GCKGFFRRTIRLKLAYDKCDRSCKIQKKNRNKCQYCRFHKCLSVGMSHNAIRFGRMPRSE
KAKLKAEILTCEHDLKDSETADLKSLAKRIHEAYLKNFNMNKVKARVILAGKTSNNPPFV
IHDMETLCMAEKTLVAKMVANGVENKEAEVRFFHCCQCMSVETVTELTEFAKAIPGFANL
DLNDQVTLLKYGVYEAIFTMLSSLMNKDGMLIAYGNGFITREFLKNLRKPFCDIMEPKFD
FAMKFNALELDDSDISLFVAAIICCGDRPGLLNIGYIEKLQEGIVHVLKLHLQSNHPDDT
FLFPKLLQKMVDLRQLVTEHAQLVQVIKKTESDAALHPLLQEIYRDMY
|
|
|
|---|
| BDBM28800 |
|---|
| BDBM10852 |
|---|
| Name | BDBM28800 |
|---|
| Synonyms: | 2-{[(3-{[2-(4-chlorophenyl)-5-methyl-1,3-oxazol-4-yl]methoxy}phenyl)methyl](methoxycarbonyl)amino}acetic acid | BMS-687453 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C22H21ClN2O6 |
|---|
| Mol. Mass. | 444.865 |
|---|
| SMILES | COC(=O)N(CC(O)=O)Cc1cccc(OCc2nc(oc2C)-c2ccc(Cl)cc2)c1 |
|---|
| Structure | 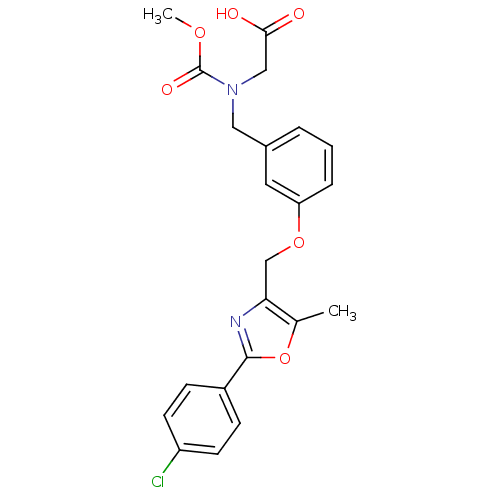
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Mukherjee, R; Locke, KT; Miao, B; Meyers, D; Monshizadegan, H; Zhang, R; Search, D; Grimm, D; Flynn, M; O'Malley, KM; Zhang, L; Li, J; Shi, Y; Kennedy, LJ; Blanar, M; Cheng, PT; Tino, J; Srivastava, RA Novel peroxisome proliferator-activated receptor alpha agonists lower low-density lipoprotein and triglycerides, raise high-density lipoprotein, and synergistically increase cholesterol excretion with a liver X receptor agonist. J Pharmacol Exp Ther327:716-26 (2008) [PubMed] Article
Mukherjee, R; Locke, KT; Miao, B; Meyers, D; Monshizadegan, H; Zhang, R; Search, D; Grimm, D; Flynn, M; O'Malley, KM; Zhang, L; Li, J; Shi, Y; Kennedy, LJ; Blanar, M; Cheng, PT; Tino, J; Srivastava, RA Novel peroxisome proliferator-activated receptor alpha agonists lower low-density lipoprotein and triglycerides, raise high-density lipoprotein, and synergistically increase cholesterol excretion with a liver X receptor agonist. J Pharmacol Exp Ther327:716-26 (2008) [PubMed] Article