| Reaction Details |
|---|
 | Report a problem with these data |
| Target | E3 ubiquitin-protein ligase XIAP |
|---|
| Ligand | BDBM13211 |
|---|
| Substrate/Competitor | BDBM32616 |
|---|
| Meas. Tech. | Fluorescence Polarization Assay |
|---|
| IC50 | 360±n/a nM |
|---|
| Citation |  Seneci, P; Bianchi, A; Battaglia, C; Belvisi, L; Bolognesi, M; Caprini, A; Cossu, F; Franco, E; Matteo, M; Delia, D; Drago, C; Khaled, A; Lecis, D; Manzoni, L; Marizzoni, M; Mastrangelo, E; Milani, M; Motto, I; Moroni, E; Potenza, D; Rizzo, V; Servida, F; Turlizzi, E; Varrone, M; Vasile, F; Scolastico, C Rational design, synthesis and characterization of potent, non-peptidic Smac mimics/XIAP inhibitors as proapoptotic agents for cancer therapy. Bioorg Med Chem17:5834-56 (2009) [PubMed] Article Seneci, P; Bianchi, A; Battaglia, C; Belvisi, L; Bolognesi, M; Caprini, A; Cossu, F; Franco, E; Matteo, M; Delia, D; Drago, C; Khaled, A; Lecis, D; Manzoni, L; Marizzoni, M; Mastrangelo, E; Milani, M; Motto, I; Moroni, E; Potenza, D; Rizzo, V; Servida, F; Turlizzi, E; Varrone, M; Vasile, F; Scolastico, C Rational design, synthesis and characterization of potent, non-peptidic Smac mimics/XIAP inhibitors as proapoptotic agents for cancer therapy. Bioorg Med Chem17:5834-56 (2009) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| E3 ubiquitin-protein ligase XIAP |
|---|
| Name: | E3 ubiquitin-protein ligase XIAP |
|---|
| Synonyms: | API3 | BIRC4 | E3 ubiquitin-protein ligase XIAP | IAP3 | Inhibitor of apoptosis protein 3 | Inhibitor of apoptosis protein 3 (XIAP) | X-linked inhibitor of apoptosis | X-linked inhibitor of apoptosis protein (XIAP) | XIAP | XIAP_HUMAN |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 56685.27 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P98170 |
|---|
| Residue: | 497 |
|---|
| Sequence: | MTFNSFEGSKTCVPADINKEEEFVEEFNRLKTFANFPSGSPVSASTLARAGFLYTGEGDT
VRCFSCHAAVDRWQYGDSAVGRHRKVSPNCRFINGFYLENSATQSTNSGIQNGQYKVENY
LGSRDHFALDRPSETHADYLLRTGQVVDISDTIYPRNPAMYSEEARLKSFQNWPDYAHLT
PRELASAGLYYTGIGDQVQCFCCGGKLKNWEPCDRAWSEHRRHFPNCFFVLGRNLNIRSE
SDAVSSDRNFPNSTNLPRNPSMADYEARIFTFGTWIYSVNKEQLARAGFYALGEGDKVKC
FHCGGGLTDWKPSEDPWEQHAKWYPGCKYLLEQKGQEYINNIHLTHSLEECLVRTTEKTP
SLTRRIDDTIFQNPMVQEAIRMGFSFKDIKKIMEEKIQISGSNYKSLEVLVADLVNAQKD
SMQDESSQTSLQKEISTEEQLRRLQEEKLCKICMDRNIAIVFVPCGHLVTCKQCAEAVDK
CPMCYTVITFKQKIFMS
|
|
|
|---|
| BDBM13211 |
|---|
| BDBM32616 |
|---|
| Name | BDBM13211 |
|---|
| Synonyms: | (3S,6S,9aS)-6-[(2S)-2-aminobutanamido]-N-(diphenylmethyl)-5-oxo-octahydro-1H-pyrrolo[1,2-a]azepine-3-carboxamide | BMC175834 Compound 1a | JMB384673 Smac001 | JMC517169 Compound 12 | conformationally constrained Smac mimetic 1 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C27H34N4O3 |
|---|
| Mol. Mass. | 462.5839 |
|---|
| SMILES | CC[C@H](N)C(=O)N[C@H]1CCC[C@H]2CC[C@H](N2C1=O)C(=O)NC(c1ccccc1)c1ccccc1 |r| |
|---|
| Structure | 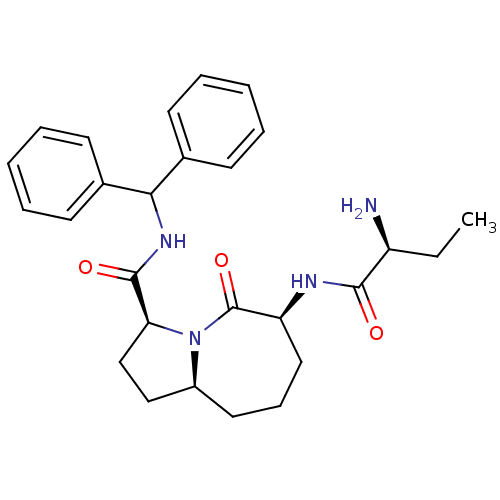
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Seneci, P; Bianchi, A; Battaglia, C; Belvisi, L; Bolognesi, M; Caprini, A; Cossu, F; Franco, E; Matteo, M; Delia, D; Drago, C; Khaled, A; Lecis, D; Manzoni, L; Marizzoni, M; Mastrangelo, E; Milani, M; Motto, I; Moroni, E; Potenza, D; Rizzo, V; Servida, F; Turlizzi, E; Varrone, M; Vasile, F; Scolastico, C Rational design, synthesis and characterization of potent, non-peptidic Smac mimics/XIAP inhibitors as proapoptotic agents for cancer therapy. Bioorg Med Chem17:5834-56 (2009) [PubMed] Article
Seneci, P; Bianchi, A; Battaglia, C; Belvisi, L; Bolognesi, M; Caprini, A; Cossu, F; Franco, E; Matteo, M; Delia, D; Drago, C; Khaled, A; Lecis, D; Manzoni, L; Marizzoni, M; Mastrangelo, E; Milani, M; Motto, I; Moroni, E; Potenza, D; Rizzo, V; Servida, F; Turlizzi, E; Varrone, M; Vasile, F; Scolastico, C Rational design, synthesis and characterization of potent, non-peptidic Smac mimics/XIAP inhibitors as proapoptotic agents for cancer therapy. Bioorg Med Chem17:5834-56 (2009) [PubMed] Article