

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Neuronal acetylcholine receptor subunit alpha-7 | ||
| Ligand | BDBM50081737 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_144339 (CHEMBL750231) | ||
| IC50 | 695±n/a nM | ||
| Citation |  Choi, KI; Cha, JH; Cho, YS; Pae, AN; Jin, C; Yook, J; Cheon, HG; Jeong, D; Kong, JY; Koh, HY Binding affinities of 3-(3-phenylisoxazol-5-yl)methylidene-1-azabicycles to acetylcholine receptors. Bioorg Med Chem Lett9:2795-800 (1999) [PubMed] Choi, KI; Cha, JH; Cho, YS; Pae, AN; Jin, C; Yook, J; Cheon, HG; Jeong, D; Kong, JY; Koh, HY Binding affinities of 3-(3-phenylisoxazol-5-yl)methylidene-1-azabicycles to acetylcholine receptors. Bioorg Med Chem Lett9:2795-800 (1999) [PubMed] | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Neuronal acetylcholine receptor subunit alpha-7 | |||
| Name: | Neuronal acetylcholine receptor subunit alpha-7 | ||
| Synonyms: | ACHA7_RAT | Acra7 | Cholinergic, Nicotinic Alpha7 | Cholinergic, Nicotinic Alpha7/5-HT3 | Chrna7 | Neuronal acetylcholine receptor | Neuronal acetylcholine receptor (alpha7 nAChR) | Neuronal acetylcholine receptor subunit alpha 7 | Neuronal acetylcholine receptor subunit alpha-7 | Neuronal acetylcholine receptor subunit alpha-7 (nAChR alpha7) | Neuronal acetylcholine receptor subunit alpha-7 (nAChR) | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 56502.44 | ||
| Organism: | Rattus norvegicus (Rat) | ||
| Description: | Q05941 | ||
| Residue: | 502 | ||
| Sequence: |
| ||
| BDBM50081737 | |||
| n/a | |||
| Name | BDBM50081737 | ||
| Synonyms: | 4-{5-[1-Aza-bicyclo[2.2.2]oct-(3E)-ylidenemethyl]-isoxazol-3-yl}-phenol | CHEMBL329782 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C17H18N2O2 | ||
| Mol. Mass. | 282.337 | ||
| SMILES | Oc1ccc(cc1)-c1cc(\C=C2\CN3CCC2CC3)on1 |(34.21,-8.55,;35.16,-9.76,;36.68,-9.57,;37.62,-10.79,;37.04,-12.21,;35.52,-12.42,;34.58,-11.19,;37.97,-13.42,;37.55,-14.88,;38.82,-15.76,;38.86,-17.31,;37.49,-18.09,;37.49,-19.61,;36.16,-20.39,;34.84,-19.61,;34.84,-18.09,;36.16,-17.31,;35.42,-19.02,;36.9,-18.61,;40.02,-14.82,;39.5,-13.38,)| | ||
| Structure | 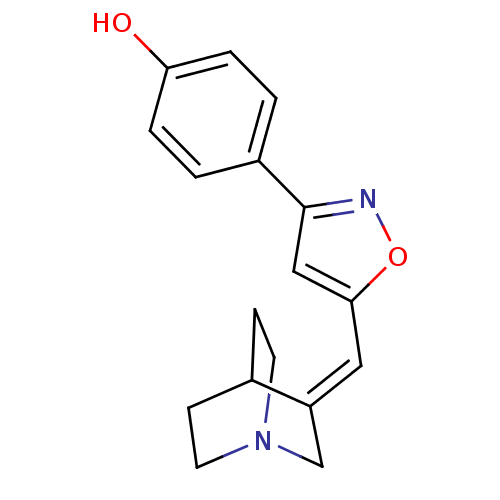
| ||