| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Indoleamine 2,3-dioxygenase 1 |
|---|
| Ligand | BDBM50578626 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_2133624 (CHEMBL4843234) |
|---|
| IC50 | 23±n/a nM |
|---|
| Citation |  Hamilton, MM; Mseeh, F; McAfoos, TJ; Leonard, PG; Reyna, NJ; Harris, AL; Xu, A; Han, M; Soth, MJ; Czako, B; Theroff, JP; Mandal, PK; Burke, JP; Virgin-Downey, B; Petrocchi, A; Pfaffinger, D; Rogers, NE; Parker, CA; Yu, SS; Jiang, Y; Krapp, S; Lammens, A; Trevitt, G; Tremblay, MR; Mikule, K; Wilcoxen, K; Cross, JB; Jones, P; Marszalek, JR; Lewis, RT Discovery of IACS-9779 and IACS-70465 as Potent Inhibitors Targeting Indoleamine 2,3-Dioxygenase 1 (IDO1) Apoenzyme. J Med Chem64:11302-11329 (2021) [PubMed] Article Hamilton, MM; Mseeh, F; McAfoos, TJ; Leonard, PG; Reyna, NJ; Harris, AL; Xu, A; Han, M; Soth, MJ; Czako, B; Theroff, JP; Mandal, PK; Burke, JP; Virgin-Downey, B; Petrocchi, A; Pfaffinger, D; Rogers, NE; Parker, CA; Yu, SS; Jiang, Y; Krapp, S; Lammens, A; Trevitt, G; Tremblay, MR; Mikule, K; Wilcoxen, K; Cross, JB; Jones, P; Marszalek, JR; Lewis, RT Discovery of IACS-9779 and IACS-70465 as Potent Inhibitors Targeting Indoleamine 2,3-Dioxygenase 1 (IDO1) Apoenzyme. J Med Chem64:11302-11329 (2021) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Indoleamine 2,3-dioxygenase 1 |
|---|
| Name: | Indoleamine 2,3-dioxygenase 1 |
|---|
| Synonyms: | I23O1_MOUSE | IDO-1 | Ido | Ido1 | Indo | Indoleamine-pyrrole 2,3-dioxygenase |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 45639.39 |
|---|
| Organism: | Mus musculus |
|---|
| Description: | ChEMBL_1452149 |
|---|
| Residue: | 407 |
|---|
| Sequence: | MALSKISPTEGSRRILEDHHIDEDVGFALPHPLVELPDAYSPWVLVARNLPVLIENGQLR
EEVEKLPTLSTDGLRGHRLQRLAHLALGYITMAYVWNRGDDDVRKVLPRNIAVPYCELSE
KLGLPPILSYADCVLANWKKKDPNGPMTYENMDILFSFPGGDCDKGFFLVSLLVEIAASP
AIKAIPTVSSAVERQDLKALEKALHDIATSLEKAKEIFKRMRDFVDPDTFFHVLRIYLSG
WKCSSKLPEGLLYEGVWDTPKMFSGGSAGQSSIFQSLDVLLGIKHEAGKESPAEFLQEMR
EYMPPAHRNFLFFLESAPPVREFVISRHNEDLTKAYNECVNGLVSVRKFHLAIVDTYIMK
PSKKKPTDGDKSEEPSNVESRGTGGTNPMTFLRSVKDTTEKALLSWP
|
|
|
|---|
| BDBM50578626 |
|---|
| n/a |
|---|
| Name | BDBM50578626 |
|---|
| Synonyms: | CHEMBL4870435 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C17H18F3N5O2 |
|---|
| Mol. Mass. | 381.3523 |
|---|
| SMILES | CC1(C)CN(CC[C@@]11NC(=O)NC1=O)c1cc(cc2cncn12)C(F)(F)F |r| |
|---|
| Structure | 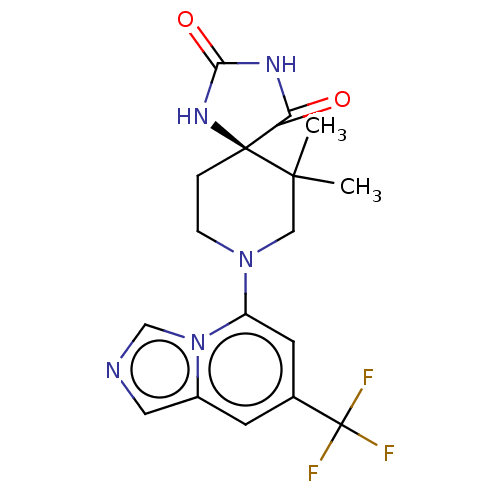
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Hamilton, MM; Mseeh, F; McAfoos, TJ; Leonard, PG; Reyna, NJ; Harris, AL; Xu, A; Han, M; Soth, MJ; Czako, B; Theroff, JP; Mandal, PK; Burke, JP; Virgin-Downey, B; Petrocchi, A; Pfaffinger, D; Rogers, NE; Parker, CA; Yu, SS; Jiang, Y; Krapp, S; Lammens, A; Trevitt, G; Tremblay, MR; Mikule, K; Wilcoxen, K; Cross, JB; Jones, P; Marszalek, JR; Lewis, RT Discovery of IACS-9779 and IACS-70465 as Potent Inhibitors Targeting Indoleamine 2,3-Dioxygenase 1 (IDO1) Apoenzyme. J Med Chem64:11302-11329 (2021) [PubMed] Article
Hamilton, MM; Mseeh, F; McAfoos, TJ; Leonard, PG; Reyna, NJ; Harris, AL; Xu, A; Han, M; Soth, MJ; Czako, B; Theroff, JP; Mandal, PK; Burke, JP; Virgin-Downey, B; Petrocchi, A; Pfaffinger, D; Rogers, NE; Parker, CA; Yu, SS; Jiang, Y; Krapp, S; Lammens, A; Trevitt, G; Tremblay, MR; Mikule, K; Wilcoxen, K; Cross, JB; Jones, P; Marszalek, JR; Lewis, RT Discovery of IACS-9779 and IACS-70465 as Potent Inhibitors Targeting Indoleamine 2,3-Dioxygenase 1 (IDO1) Apoenzyme. J Med Chem64:11302-11329 (2021) [PubMed] Article