| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Peptide deformylase |
|---|
| Ligand | BDBM50255002 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_559976 (CHEMBL1014575) |
|---|
| IC50 | 15±n/a nM |
|---|
| Citation |  Pichota, A; Duraiswamy, J; Yin, Z; Keller, TH; Alam, J; Liung, S; Lee, G; Ding, M; Wang, G; Chan, WL; Schreiber, M; Ma, I; Beer, D; Ngew, X; Mukherjee, K; Nanjundappa, M; Teo, JW; Thayalan, P; Yap, A; Dick, T; Meng, W; Xu, M; Koehn, J; Pan, SH; Clark, K; Xie, X; Shoen, C; Cynamon, M Peptide deformylase inhibitors of Mycobacterium tuberculosis: synthesis, structural investigations, and biological results. Bioorg Med Chem Lett18:6568-72 (2008) [PubMed] Article Pichota, A; Duraiswamy, J; Yin, Z; Keller, TH; Alam, J; Liung, S; Lee, G; Ding, M; Wang, G; Chan, WL; Schreiber, M; Ma, I; Beer, D; Ngew, X; Mukherjee, K; Nanjundappa, M; Teo, JW; Thayalan, P; Yap, A; Dick, T; Meng, W; Xu, M; Koehn, J; Pan, SH; Clark, K; Xie, X; Shoen, C; Cynamon, M Peptide deformylase inhibitors of Mycobacterium tuberculosis: synthesis, structural investigations, and biological results. Bioorg Med Chem Lett18:6568-72 (2008) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Peptide deformylase |
|---|
| Name: | Peptide deformylase |
|---|
| Synonyms: | DEF_MYCTU | def |
|---|
| Type: | PROTEIN |
|---|
| Mol. Mass.: | 20931.95 |
|---|
| Organism: | Mycobacterium tuberculosis |
|---|
| Description: | ChEMBL_559976 |
|---|
| Residue: | 197 |
|---|
| Sequence: | MAVVPIRIVGDPVLHTATTPVTVAADGSLPADLAQLIATMYDTMDAANGVGLAANQIGCS
LRLFVYDCAADRAMTARRRGVVINPVLETSEIPETMPDPDTDDEGCLSVPGESFPTGRAK
WARVTGLDADGSPVSIEGTGLFARMLQHETGHLDGFLYLDRLIGRYARNAKRAVKSHGWG
VPGLSWLPGEDPDPFGH
|
|
|
|---|
| BDBM50255002 |
|---|
| n/a |
|---|
| Name | BDBM50255002 |
|---|
| Synonyms: | CHEMBL482180 | N-((R)-2-((S)-2-(1H-benzo[d]imidazol-2-yl)pyrrolidine-1-carbonyl)heptyl)-N-hydroxyformamide |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C20H28N4O3 |
|---|
| Mol. Mass. | 372.4613 |
|---|
| SMILES | CCCCC[C@H](CN(O)C=O)C(=O)N1CCC[C@H]1c1nc2ccccc2[nH]1 |r| |
|---|
| Structure | 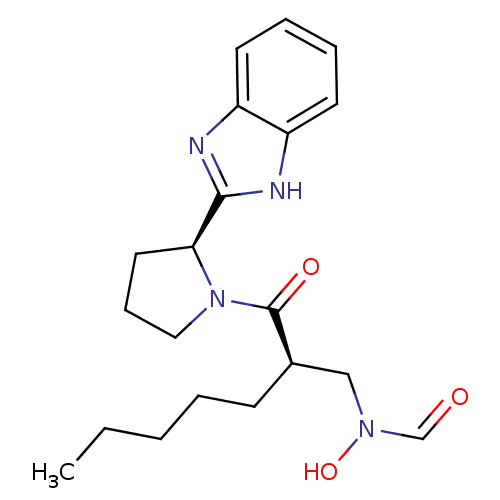
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Pichota, A; Duraiswamy, J; Yin, Z; Keller, TH; Alam, J; Liung, S; Lee, G; Ding, M; Wang, G; Chan, WL; Schreiber, M; Ma, I; Beer, D; Ngew, X; Mukherjee, K; Nanjundappa, M; Teo, JW; Thayalan, P; Yap, A; Dick, T; Meng, W; Xu, M; Koehn, J; Pan, SH; Clark, K; Xie, X; Shoen, C; Cynamon, M Peptide deformylase inhibitors of Mycobacterium tuberculosis: synthesis, structural investigations, and biological results. Bioorg Med Chem Lett18:6568-72 (2008) [PubMed] Article
Pichota, A; Duraiswamy, J; Yin, Z; Keller, TH; Alam, J; Liung, S; Lee, G; Ding, M; Wang, G; Chan, WL; Schreiber, M; Ma, I; Beer, D; Ngew, X; Mukherjee, K; Nanjundappa, M; Teo, JW; Thayalan, P; Yap, A; Dick, T; Meng, W; Xu, M; Koehn, J; Pan, SH; Clark, K; Xie, X; Shoen, C; Cynamon, M Peptide deformylase inhibitors of Mycobacterium tuberculosis: synthesis, structural investigations, and biological results. Bioorg Med Chem Lett18:6568-72 (2008) [PubMed] Article