| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Peroxisome proliferator-activated receptor delta |
|---|
| Ligand | BDBM50257254 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_562738 (CHEMBL1022410) |
|---|
| EC50 | >10000±n/a nM |
|---|
| Citation |  Lin, CH; Peng, YH; Coumar, MS; Chittimalla, SK; Liao, CC; Lyn, PC; Huang, CC; Lien, TW; Lin, WH; Hsu, JT; Cheng, JH; Chen, X; Wu, JS; Chao, YS; Lee, HJ; Juo, CG; Wu, SY; Hsieh, HP Design and structural analysis of novel pharmacophores for potent and selective peroxisome proliferator-activated receptor gamma agonists. J Med Chem52:2618-22 (2009) [PubMed] Article Lin, CH; Peng, YH; Coumar, MS; Chittimalla, SK; Liao, CC; Lyn, PC; Huang, CC; Lien, TW; Lin, WH; Hsu, JT; Cheng, JH; Chen, X; Wu, JS; Chao, YS; Lee, HJ; Juo, CG; Wu, SY; Hsieh, HP Design and structural analysis of novel pharmacophores for potent and selective peroxisome proliferator-activated receptor gamma agonists. J Med Chem52:2618-22 (2009) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Peroxisome proliferator-activated receptor delta |
|---|
| Name: | Peroxisome proliferator-activated receptor delta |
|---|
| Synonyms: | NR1C2 | NUC1 | NUCI | Nuclear hormone receptor 1 | Nuclear receptor subfamily 1 group C member 2 | PPAR delta | PPAR-beta | PPARB | PPARD | PPARD_HUMAN | Peroxisome proliferator-activated receptor | Peroxisome proliferator-activated receptor beta | Peroxisome proliferator-activated receptor delta |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 49910.45 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q03181 |
|---|
| Residue: | 441 |
|---|
| Sequence: | MEQPQEEAPEVREEEEKEEVAEAEGAPELNGGPQHALPSSSYTDLSRSSSPPSLLDQLQM
GCDGASCGSLNMECRVCGDKASGFHYGVHACEGCKGFFRRTIRMKLEYEKCERSCKIQKK
NRNKCQYCRFQKCLALGMSHNAIRFGRMPEAEKRKLVAGLTANEGSQYNPQVADLKAFSK
HIYNAYLKNFNMTKKKARSILTGKASHTAPFVIHDIETLWQAEKGLVWKQLVNGLPPYKE
ISVHVFYRCQCTTVETVRELTEFAKSIPSFSSLFLNDQVTLLKYGVHEAIFAMLASIVNK
DGLLVANGSGFVTREFLRSLRKPFSDIIEPKFEFAVKFNALELDDSDLALFIAAIILCGD
RPGLMNVPRVEAIQDTILRALEFHLQANHPDAQYLFPKLLQKMADLRQLVTEHAQMMQRI
KKTETETSLHPLLQEIYKDMY
|
|
|
|---|
| BDBM50257254 |
|---|
| n/a |
|---|
| Name | BDBM50257254 |
|---|
| Synonyms: | 2-{1-[3-(5-Benzoyl-1-propyl-naphthalen-2-yloxy)-propyl]-1H-indol-5-yloxy}-2-methyl-propionicacid | CHEMBL454508 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C35H35NO5 |
|---|
| Mol. Mass. | 549.6561 |
|---|
| SMILES | CCCc1c(OCCCn2ccc3cc(OC(C)(C)C(O)=O)ccc23)ccc2c(cccc12)C(=O)c1ccccc1 |
|---|
| Structure | 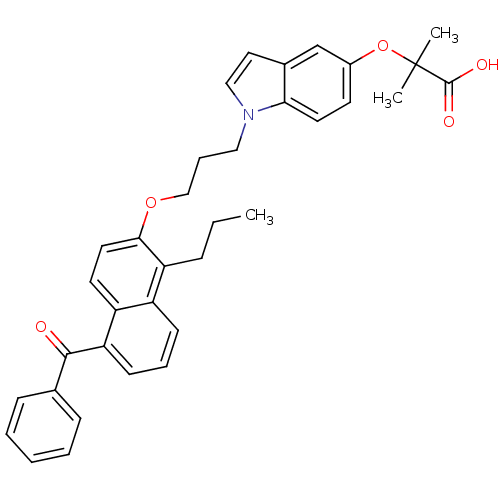
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Lin, CH; Peng, YH; Coumar, MS; Chittimalla, SK; Liao, CC; Lyn, PC; Huang, CC; Lien, TW; Lin, WH; Hsu, JT; Cheng, JH; Chen, X; Wu, JS; Chao, YS; Lee, HJ; Juo, CG; Wu, SY; Hsieh, HP Design and structural analysis of novel pharmacophores for potent and selective peroxisome proliferator-activated receptor gamma agonists. J Med Chem52:2618-22 (2009) [PubMed] Article
Lin, CH; Peng, YH; Coumar, MS; Chittimalla, SK; Liao, CC; Lyn, PC; Huang, CC; Lien, TW; Lin, WH; Hsu, JT; Cheng, JH; Chen, X; Wu, JS; Chao, YS; Lee, HJ; Juo, CG; Wu, SY; Hsieh, HP Design and structural analysis of novel pharmacophores for potent and selective peroxisome proliferator-activated receptor gamma agonists. J Med Chem52:2618-22 (2009) [PubMed] Article