| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Tyrosine-protein kinase ABL1 |
|---|
| Ligand | BDBM50322535 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_641699 (CHEMBL1176866) |
|---|
| IC50 | 0.37±n/a nM |
|---|
| Citation |  Huang, WS; Metcalf, CA; Sundaramoorthi, R; Wang, Y; Zou, D; Thomas, RM; Zhu, X; Cai, L; Wen, D; Liu, S; Romero, J; Qi, J; Chen, I; Banda, G; Lentini, SP; Das, S; Xu, Q; Keats, J; Wang, F; Wardwell, S; Ning, Y; Snodgrass, JT; Broudy, MI; Russian, K; Zhou, T; Commodore, L; Narasimhan, NI; Mohemmad, QK; Iuliucci, J; Rivera, VM; Dalgarno, DC; Sawyer, TK; Clackson, T; Shakespeare, WC Discovery of 3-[2-(imidazo[1,2-b]pyridazin-3-yl)ethynyl]-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzamide (AP24534), a potent, orally active pan-inhibitor of breakpoint cluster region-abelson (BCR-ABL) kinase including the T315I gatekeeper mutant. J Med Chem53:4701-19 (2010) [PubMed] Article Huang, WS; Metcalf, CA; Sundaramoorthi, R; Wang, Y; Zou, D; Thomas, RM; Zhu, X; Cai, L; Wen, D; Liu, S; Romero, J; Qi, J; Chen, I; Banda, G; Lentini, SP; Das, S; Xu, Q; Keats, J; Wang, F; Wardwell, S; Ning, Y; Snodgrass, JT; Broudy, MI; Russian, K; Zhou, T; Commodore, L; Narasimhan, NI; Mohemmad, QK; Iuliucci, J; Rivera, VM; Dalgarno, DC; Sawyer, TK; Clackson, T; Shakespeare, WC Discovery of 3-[2-(imidazo[1,2-b]pyridazin-3-yl)ethynyl]-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzamide (AP24534), a potent, orally active pan-inhibitor of breakpoint cluster region-abelson (BCR-ABL) kinase including the T315I gatekeeper mutant. J Med Chem53:4701-19 (2010) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Tyrosine-protein kinase ABL1 |
|---|
| Name: | Tyrosine-protein kinase ABL1 |
|---|
| Synonyms: | ABL | ABL1 | ABL1_HUMAN | Abelson murine leukemia viral oncogene homolog 1 | JTK7 | Proto-oncogene c-Abl | Proto-oncogene tyrosine-protein kinase ABL1 | Tyrosine-protein kinase (ABL) | Tyrosine-protein kinase ABL | Tyrosine-protein kinase ABL1 (ABL) | V-abl Abelson murine leukemia viral oncogene homolog 1 | c-ABL | p150 | tyrosine-protein kinase ABL1 isoform a |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 122897.30 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P00519 |
|---|
| Residue: | 1130 |
|---|
| Sequence: | MLEICLKLVGCKSKKGLSSSSSCYLEEALQRPVASDFEPQGLSEAARWNSKENLLAGPSE
NDPNLFVALYDFVASGDNTLSITKGEKLRVLGYNHNGEWCEAQTKNGQGWVPSNYITPVN
SLEKHSWYHGPVSRNAAEYLLSSGINGSFLVRESESSPGQRSISLRYEGRVYHYRINTAS
DGKLYVSSESRFNTLAELVHHHSTVADGLITTLHYPAPKRNKPTVYGVSPNYDKWEMERT
DITMKHKLGGGQYGEVYEGVWKKYSLTVAVKTLKEDTMEVEEFLKEAAVMKEIKHPNLVQ
LLGVCTREPPFYIITEFMTYGNLLDYLRECNRQEVNAVVLLYMATQISSAMEYLEKKNFI
HRDLAARNCLVGENHLVKVADFGLSRLMTGDTYTAHAGAKFPIKWTAPESLAYNKFSIKS
DVWAFGVLLWEIATYGMSPYPGIDLSQVYELLEKDYRMERPEGCPEKVYELMRACWQWNP
SDRPSFAEIHQAFETMFQESSISDEVEKELGKQGVRGAVSTLLQAPELPTKTRTSRRAAE
HRDTTDVPEMPHSKGQGESDPLDHEPAVSPLLPRKERGPPEGGLNEDERLLPKDKKTNLF
SALIKKKKKTAPTPPKRSSSFREMDGQPERRGAGEEEGRDISNGALAFTPLDTADPAKSP
KPSNGAGVPNGALRESGGSGFRSPHLWKKSSTLTSSRLATGEEEGGGSSSKRFLRSCSAS
CVPHGAKDTEWRSVTLPRDLQSTGRQFDSSTFGGHKSEKPALPRKRAGENRSDQVTRGTV
TPPPRLVKKNEEAADEVFKDIMESSPGSSPPNLTPKPLRRQVTVAPASGLPHKEEAGKGS
ALGTPAAAEPVTPTSKAGSGAPGGTSKGPAEESRVRRHKHSSESPGRDKGKLSRLKPAPP
PPPAASAGKAGGKPSQSPSQEAAGEAVLGAKTKATSLVDAVNSDAAKPSQPGEGLKKPVL
PATPKPQSAKPSGTPISPAPVPSTLPSASSALAGDQPSSTAFIPLISTRVSLRKTRQPPE
RIASGAITKGVVLDSTEALCLAISRNSEQMASHSAVLEAGKNLYTFCVSYVDSIQQMRNK
FAFREAINKLENNLRELQICPATAGSGPAATQDFSKLLSSVKEISDIVQR
|
|
|
|---|
| BDBM50322535 |
|---|
| n/a |
|---|
| Name | BDBM50322535 |
|---|
| Synonyms: | 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzamide | 3-[2-(Imidazo[1,2-b]pyridazin-3-yl)ethynyl]-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzamide | CHEMBL1171837 | PONATINIB | US10464902, Ponatinib | US9255107, AP24534 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C29H27F3N6O |
|---|
| Mol. Mass. | 532.5595 |
|---|
| SMILES | CN1CCN(Cc2ccc(NC(=O)c3ccc(C)c(c3)C#Cc3cnc4cccnn34)cc2C(F)(F)F)CC1 |
|---|
| Structure | 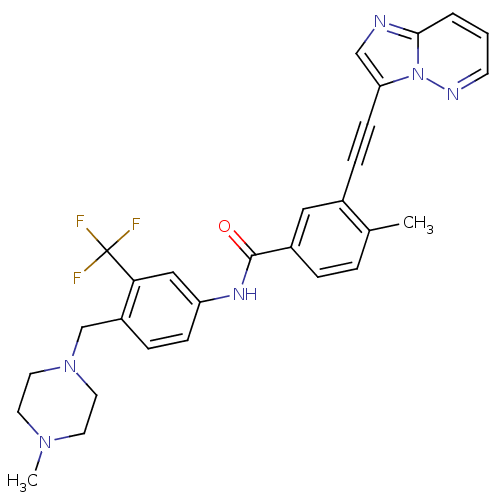
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Huang, WS; Metcalf, CA; Sundaramoorthi, R; Wang, Y; Zou, D; Thomas, RM; Zhu, X; Cai, L; Wen, D; Liu, S; Romero, J; Qi, J; Chen, I; Banda, G; Lentini, SP; Das, S; Xu, Q; Keats, J; Wang, F; Wardwell, S; Ning, Y; Snodgrass, JT; Broudy, MI; Russian, K; Zhou, T; Commodore, L; Narasimhan, NI; Mohemmad, QK; Iuliucci, J; Rivera, VM; Dalgarno, DC; Sawyer, TK; Clackson, T; Shakespeare, WC Discovery of 3-[2-(imidazo[1,2-b]pyridazin-3-yl)ethynyl]-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzamide (AP24534), a potent, orally active pan-inhibitor of breakpoint cluster region-abelson (BCR-ABL) kinase including the T315I gatekeeper mutant. J Med Chem53:4701-19 (2010) [PubMed] Article
Huang, WS; Metcalf, CA; Sundaramoorthi, R; Wang, Y; Zou, D; Thomas, RM; Zhu, X; Cai, L; Wen, D; Liu, S; Romero, J; Qi, J; Chen, I; Banda, G; Lentini, SP; Das, S; Xu, Q; Keats, J; Wang, F; Wardwell, S; Ning, Y; Snodgrass, JT; Broudy, MI; Russian, K; Zhou, T; Commodore, L; Narasimhan, NI; Mohemmad, QK; Iuliucci, J; Rivera, VM; Dalgarno, DC; Sawyer, TK; Clackson, T; Shakespeare, WC Discovery of 3-[2-(imidazo[1,2-b]pyridazin-3-yl)ethynyl]-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzamide (AP24534), a potent, orally active pan-inhibitor of breakpoint cluster region-abelson (BCR-ABL) kinase including the T315I gatekeeper mutant. J Med Chem53:4701-19 (2010) [PubMed] Article