

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Insulin-like growth factor 1 receptor | ||
| Ligand | BDBM50219382 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_716595 (CHEMBL1670374) | ||
| IC50 | 19±n/a nM | ||
| Citation |  Jin, M; Kleinberg, A; Cooke, A; Gokhale, PC; Foreman, K; Dong, H; Siu, KW; Bittner, MA; Mulvihill, KM; Yao, Y; Landfair, D; O'Connor, M; Mak, G; Pachter, JA; Wild, R; Rosenfeld-Franklin, M; Ji, Q; Mulvihill, MJ Potent and selective cyclohexyl-derived imidazopyrazine insulin-like growth factor 1 receptor inhibitors with in vivo efficacy. Bioorg Med Chem Lett21:1176-80 (2011) [PubMed] Article Jin, M; Kleinberg, A; Cooke, A; Gokhale, PC; Foreman, K; Dong, H; Siu, KW; Bittner, MA; Mulvihill, KM; Yao, Y; Landfair, D; O'Connor, M; Mak, G; Pachter, JA; Wild, R; Rosenfeld-Franklin, M; Ji, Q; Mulvihill, MJ Potent and selective cyclohexyl-derived imidazopyrazine insulin-like growth factor 1 receptor inhibitors with in vivo efficacy. Bioorg Med Chem Lett21:1176-80 (2011) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Insulin-like growth factor 1 receptor | |||
| Name: | Insulin-like growth factor 1 receptor | ||
| Synonyms: | CD_antigen=CD221 | IGF-I receptor | IGF1R | IGF1R_HUMAN | Insulin-like growth factor 1 receptor (IGF1R) | Insulin-like growth factor 1 receptor (IGFIR) | Insulin-like growth factor 1 receptor alpha chain | Insulin-like growth factor 1 receptor beta chain | Insulin-like growth factor I receptor | Insulin-like growth factor receptor (IGFR) | ||
| Type: | Protein | ||
| Mol. Mass.: | 154776.79 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | P08069 | ||
| Residue: | 1367 | ||
| Sequence: |
| ||
| BDBM50219382 | |||
| n/a | |||
| Name | BDBM50219382 | ||
| Synonyms: | 3-((1s,3s)-3-(4-methylpiperazin-1-yl)cyclobutyl)-1-(2-phenylquinolin-7-yl)imidazo[1,5-a]pyrazin-8-amine | 3-[cis-3-(4-methylpiperazin-1-yl)cyclobutyl]-1-(2-phenylquinolin-7-yl)imidazo[1,5-a]pyrazin-8-amine | CHEMBL249295 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C30H31N7 | ||
| Mol. Mass. | 489.614 | ||
| SMILES | CN1CCN(CC1)[C@H]1C[C@H](C1)c1nc(-c2ccc3ccc(nc3c2)-c2ccccc2)c2c(N)nccn12 |wU:7.7,9.12,(13.72,-11.86,;13.25,-10.4,;11.74,-10.07,;11.27,-8.61,;12.31,-7.47,;13.81,-7.78,;14.28,-9.25,;11.83,-6.01,;10.46,-5.31,;11.15,-3.94,;12.53,-4.64,;10.68,-2.48,;11.59,-1.22,;10.67,.03,;11.15,1.5,;10.12,2.63,;10.58,4.09,;12.09,4.42,;12.57,5.88,;14.08,6.21,;15.12,5.05,;14.63,3.58,;13.12,3.27,;12.65,1.81,;16.62,5.37,;17.65,4.22,;19.15,4.54,;19.63,6.01,;18.59,7.15,;17.09,6.83,;9.2,-.45,;7.86,.32,;7.85,1.86,;6.53,-.45,;6.53,-2,;7.86,-2.77,;9.2,-2,)| | ||
| Structure | 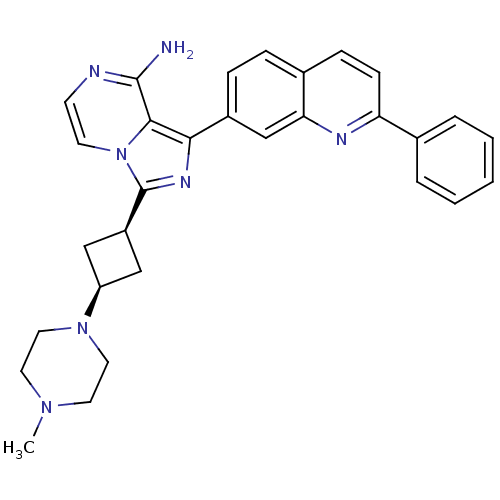
| ||