| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Mitogen-activated protein kinase 1 |
|---|
| Ligand | BDBM50182212 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_346249 (CHEMBL861204) |
|---|
| IC50 | 12600±n/a nM |
|---|
| Citation |  McDermott, LA; Higgins, B; Simcox, M; Luk, KC; Nevins, T; Kolinsky, K; Smith, M; Yang, H; Li, JK; Chen, Y; Ke, J; Mallalieu, N; Egan, T; Kolis, S; Railkar, A; Gerber, L; Liu, JJ; Konzelmann, F; Zhang, Z; Flynn, T; Morales, O; Chen, Y Biological evaluation of a multi-targeted small molecule inhibitor of tumor-induced angiogenesis. Bioorg Med Chem Lett16:1950-3 (2006) [PubMed] Article McDermott, LA; Higgins, B; Simcox, M; Luk, KC; Nevins, T; Kolinsky, K; Smith, M; Yang, H; Li, JK; Chen, Y; Ke, J; Mallalieu, N; Egan, T; Kolis, S; Railkar, A; Gerber, L; Liu, JJ; Konzelmann, F; Zhang, Z; Flynn, T; Morales, O; Chen, Y Biological evaluation of a multi-targeted small molecule inhibitor of tumor-induced angiogenesis. Bioorg Med Chem Lett16:1950-3 (2006) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Mitogen-activated protein kinase 1 |
|---|
| Name: | Mitogen-activated protein kinase 1 |
|---|
| Synonyms: | ERK2 | ERT1 | Extracellular signal-regulated kinase 2 | Extracellular signal-regulated kinase 2 (ERK-2) | Extracellular signal-regulated kinase 2 (ERK2) | MAP Kinase 2/ERK2 | MAPK 2 | MAPK1 | MK01_HUMAN | Mitogen activated kinase 1 (ERK-2) | Mitogen-activated protein kinase 1 (ERK-2) | Mitogen-activated protein kinase 1 (ERK2) | Mitogen-activated protein kinase 2 | PRKM1 | PRKM2 | p42-MAPK |
|---|
| Type: | Ser/Thr Protein Kinase |
|---|
| Mol. Mass.: | 41392.76 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P28482 |
|---|
| Residue: | 360 |
|---|
| Sequence: | MAAAAAAGAGPEMVRGQVFDVGPRYTNLSYIGEGAYGMVCSAYDNVNKVRVAIKKISPFE
HQTYCQRTLREIKILLRFRHENIIGINDIIRAPTIEQMKDVYIVQDLMETDLYKLLKTQH
LSNDHICYFLYQILRGLKYIHSANVLHRDLKPSNLLLNTTCDLKICDFGLARVADPDHDH
TGFLTEYVATRWYRAPEIMLNSKGYTKSIDIWSVGCILAEMLSNRPIFPGKHYLDQLNHI
LGILGSPSQEDLNCIINLKARNYLLSLPHKNKVPWNRLFPNADSKALDLLDKMLTFNPHK
RIEVEQALAHPYLEQYYDPSDEPIAEAPFKFDMELDDLPKEKLKELIFEETARFQPGYRS
|
|
|
|---|
| BDBM50182212 |
|---|
| n/a |
|---|
| Name | BDBM50182212 |
|---|
| Synonyms: | 7-(4-fluorophenylamino)-1-((1S,3R)-3-hydroxycyclopentyl)-3-(4-methoxyphenyl)-3,4-dihydropyrimido[4,5-d]pyrimidin-2(1H)-one |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C24H24FN5O3 |
|---|
| Mol. Mass. | 449.4775 |
|---|
| SMILES | COc1ccc(cc1)N1Cc2cnc(Nc3ccc(F)cc3)nc2N(C2CC[C@@H](O)C2)C1=O |
|---|
| Structure | 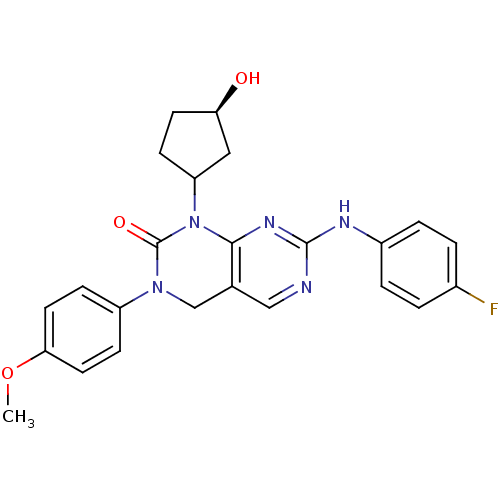
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 McDermott, LA; Higgins, B; Simcox, M; Luk, KC; Nevins, T; Kolinsky, K; Smith, M; Yang, H; Li, JK; Chen, Y; Ke, J; Mallalieu, N; Egan, T; Kolis, S; Railkar, A; Gerber, L; Liu, JJ; Konzelmann, F; Zhang, Z; Flynn, T; Morales, O; Chen, Y Biological evaluation of a multi-targeted small molecule inhibitor of tumor-induced angiogenesis. Bioorg Med Chem Lett16:1950-3 (2006) [PubMed] Article
McDermott, LA; Higgins, B; Simcox, M; Luk, KC; Nevins, T; Kolinsky, K; Smith, M; Yang, H; Li, JK; Chen, Y; Ke, J; Mallalieu, N; Egan, T; Kolis, S; Railkar, A; Gerber, L; Liu, JJ; Konzelmann, F; Zhang, Z; Flynn, T; Morales, O; Chen, Y Biological evaluation of a multi-targeted small molecule inhibitor of tumor-induced angiogenesis. Bioorg Med Chem Lett16:1950-3 (2006) [PubMed] Article