| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Beta-secretase 1 |
|---|
| Ligand | BDBM50323511 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_644781 (CHEMBL1211024) |
|---|
| IC50 | 100±n/a nM |
|---|
| Citation |  Truong, AP; Probst, GD; Aquino, J; Fang, L; Brogley, L; Sealy, JM; Hom, RK; Tucker, JA; John, V; Tung, JS; Pleiss, MA; Konradi, AW; Sham, HL; Dappen, MS; Tóth, G; Yao, N; Brecht, E; Pan, H; Artis, DR; Ruslim, L; Bova, MP; Sinha, S; Yednock, TA; Zmolek, W; Quinn, KP; Sauer, JM Improving the permeability of the hydroxyethylamine BACE-1 inhibitors: structure-activity relationship of P2' substituents. Bioorg Med Chem Lett20:4789-94 (2010) [PubMed] Article Truong, AP; Probst, GD; Aquino, J; Fang, L; Brogley, L; Sealy, JM; Hom, RK; Tucker, JA; John, V; Tung, JS; Pleiss, MA; Konradi, AW; Sham, HL; Dappen, MS; Tóth, G; Yao, N; Brecht, E; Pan, H; Artis, DR; Ruslim, L; Bova, MP; Sinha, S; Yednock, TA; Zmolek, W; Quinn, KP; Sauer, JM Improving the permeability of the hydroxyethylamine BACE-1 inhibitors: structure-activity relationship of P2' substituents. Bioorg Med Chem Lett20:4789-94 (2010) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Beta-secretase 1 |
|---|
| Name: | Beta-secretase 1 |
|---|
| Synonyms: | ASP2 | Asp 2 | Aspartyl protease 2 | BACE | BACE1 | BACE1_HUMAN | Beta-secretase (BACE) | Beta-secretase 1 | Beta-secretase 1 (BACE 1) | Beta-secretase 1 (BACE-1) | Beta-secretase 1 (BACE1) | Beta-site APP cleaving enzyme 1 | Beta-site amyloid precursor protein cleaving enzyme 1 | KIAA1149 | Memapsin-2 | Membrane-associated aspartic protease 2 | beta-Secretase (BACE-1) | beta-Secretase (BACE1) |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 55755.10 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | P56817 |
|---|
| Residue: | 501 |
|---|
| Sequence: | MAQALPWLLLWMGAGVLPAHGTQHGIRLPLRSGLGGAPLGLRLPRETDEEPEEPGRRGSF
VEMVDNLRGKSGQGYYVEMTVGSPPQTLNILVDTGSSNFAVGAAPHPFLHRYYQRQLSST
YRDLRKGVYVPYTQGKWEGELGTDLVSIPHGPNVTVRANIAAITESDKFFINGSNWEGIL
GLAYAEIARPDDSLEPFFDSLVKQTHVPNLFSLQLCGAGFPLNQSEVLASVGGSMIIGGI
DHSLYTGSLWYTPIRREWYYEVIIVRVEINGQDLKMDCKEYNYDKSIVDSGTTNLRLPKK
VFEAAVKSIKAASSTEKFPDGFWLGEQLVCWQAGTTPWNIFPVISLYLMGEVTNQSFRIT
ILPQQYLRPVEDVATSQDDCYKFAISQSSTGTVMGAVIMEGFYVVFDRARKRIGFAVSAC
HVHDEFRTAAVEGPFVTLDMEDCGYNIPQTDESTLMTIAYVMAAICALFMLPLCLMVCQW
RCLRCLRQQHDDFADDISLLK
|
|
|
|---|
| BDBM50323511 |
|---|
| n/a |
|---|
| Name | BDBM50323511 |
|---|
| Synonyms: | CHEMBL1209894 | N-((2S,3R)-1-(3,5-difluorophenyl)-3-hydroxy-4-(1-(3-(prop-1-en-2-yl)phenyl)cyclohexylamino)butan-2-yl)acetamide |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C27H34F2N2O2 |
|---|
| Mol. Mass. | 456.5679 |
|---|
| SMILES | CC(=O)N[C@@H](Cc1cc(F)cc(F)c1)[C@H](O)CNC1(CCCCC1)c1cccc(c1)C(C)=C |r| |
|---|
| Structure | 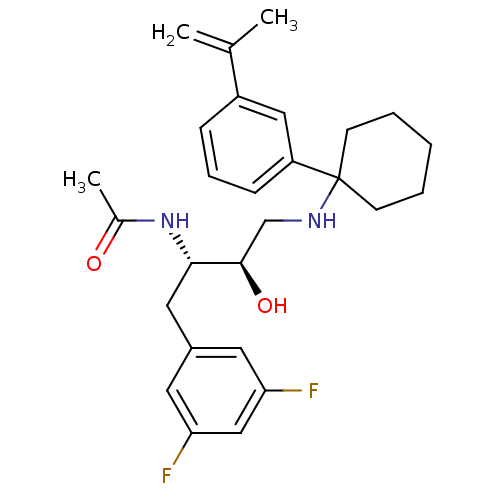
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Truong, AP; Probst, GD; Aquino, J; Fang, L; Brogley, L; Sealy, JM; Hom, RK; Tucker, JA; John, V; Tung, JS; Pleiss, MA; Konradi, AW; Sham, HL; Dappen, MS; Tóth, G; Yao, N; Brecht, E; Pan, H; Artis, DR; Ruslim, L; Bova, MP; Sinha, S; Yednock, TA; Zmolek, W; Quinn, KP; Sauer, JM Improving the permeability of the hydroxyethylamine BACE-1 inhibitors: structure-activity relationship of P2' substituents. Bioorg Med Chem Lett20:4789-94 (2010) [PubMed] Article
Truong, AP; Probst, GD; Aquino, J; Fang, L; Brogley, L; Sealy, JM; Hom, RK; Tucker, JA; John, V; Tung, JS; Pleiss, MA; Konradi, AW; Sham, HL; Dappen, MS; Tóth, G; Yao, N; Brecht, E; Pan, H; Artis, DR; Ruslim, L; Bova, MP; Sinha, S; Yednock, TA; Zmolek, W; Quinn, KP; Sauer, JM Improving the permeability of the hydroxyethylamine BACE-1 inhibitors: structure-activity relationship of P2' substituents. Bioorg Med Chem Lett20:4789-94 (2010) [PubMed] Article