| Citation |  Cook, J; Zusi, FC; McDonald, IM; King, D; Hill, MD; Iwuagwu, C; Mate, RA; Fang, H; Zhao, R; Wang, B; Cutrone, J; Ma, B; Gao, Q; Knox, RJ; Matchett, M; Gallagher, L; Ferrante, M; Post-Munson, D; Molski, T; Easton, A; Miller, R; Jones, K; Digavalli, S; Healy, F; Lentz, K; Benitex, Y; Clarke, W; Natale, J; Siuciak, JA; Lodge, N; Zaczek, R; Denton, R; Morgan, D; Bristow, LJ; Macor, JE; Olson, RE Design and Synthesis of a New Series of 4-Heteroarylamino-1'-azaspiro[oxazole-5,3'-bicyclo[2.2.2]octanes asa7 Nicotinic Receptor Agonists. 1. Development of Pharmacophore and Early Structure-Activity Relationship. J Med Chem59:11171-11181 (2016) [PubMed] Article Cook, J; Zusi, FC; McDonald, IM; King, D; Hill, MD; Iwuagwu, C; Mate, RA; Fang, H; Zhao, R; Wang, B; Cutrone, J; Ma, B; Gao, Q; Knox, RJ; Matchett, M; Gallagher, L; Ferrante, M; Post-Munson, D; Molski, T; Easton, A; Miller, R; Jones, K; Digavalli, S; Healy, F; Lentz, K; Benitex, Y; Clarke, W; Natale, J; Siuciak, JA; Lodge, N; Zaczek, R; Denton, R; Morgan, D; Bristow, LJ; Macor, JE; Olson, RE Design and Synthesis of a New Series of 4-Heteroarylamino-1'-azaspiro[oxazole-5,3'-bicyclo[2.2.2]octanes asa7 Nicotinic Receptor Agonists. 1. Development of Pharmacophore and Early Structure-Activity Relationship. J Med Chem59:11171-11181 (2016) [PubMed] Article |
|---|
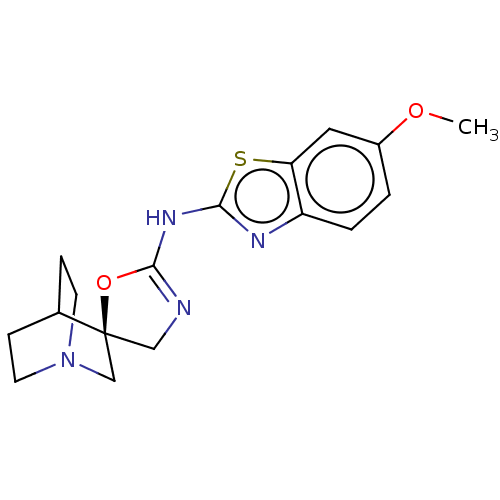
| SMILES | COc1ccc2nc(NC3=NC[C@@]4(CN5CCC4CC5)O3)sc2c1 |r,wU:12.21,t:9,THB:20:12:15.16:19.18,(37.09,-40.85,;35.6,-40.46,;34.52,-41.55,;34.92,-43.04,;33.83,-44.12,;32.35,-43.73,;31.07,-44.57,;29.87,-43.61,;28.38,-44.01,;27.98,-45.5,;28.96,-46.69,;28.12,-47.99,;26.63,-47.59,;26.93,-49.11,;25.44,-48.42,;23.78,-49.14,;23.56,-47.64,;25.16,-46.94,;25.24,-45.16,;25.72,-46.36,;26.55,-46.05,;30.42,-42.17,;31.96,-42.24,;33.04,-41.16,)| |
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Cook, J; Zusi, FC; McDonald, IM; King, D; Hill, MD; Iwuagwu, C; Mate, RA; Fang, H; Zhao, R; Wang, B; Cutrone, J; Ma, B; Gao, Q; Knox, RJ; Matchett, M; Gallagher, L; Ferrante, M; Post-Munson, D; Molski, T; Easton, A; Miller, R; Jones, K; Digavalli, S; Healy, F; Lentz, K; Benitex, Y; Clarke, W; Natale, J; Siuciak, JA; Lodge, N; Zaczek, R; Denton, R; Morgan, D; Bristow, LJ; Macor, JE; Olson, RE Design and Synthesis of a New Series of 4-Heteroarylamino-1'-azaspiro[oxazole-5,3'-bicyclo[2.2.2]octanes asa7 Nicotinic Receptor Agonists. 1. Development of Pharmacophore and Early Structure-Activity Relationship. J Med Chem59:11171-11181 (2016) [PubMed] Article
Cook, J; Zusi, FC; McDonald, IM; King, D; Hill, MD; Iwuagwu, C; Mate, RA; Fang, H; Zhao, R; Wang, B; Cutrone, J; Ma, B; Gao, Q; Knox, RJ; Matchett, M; Gallagher, L; Ferrante, M; Post-Munson, D; Molski, T; Easton, A; Miller, R; Jones, K; Digavalli, S; Healy, F; Lentz, K; Benitex, Y; Clarke, W; Natale, J; Siuciak, JA; Lodge, N; Zaczek, R; Denton, R; Morgan, D; Bristow, LJ; Macor, JE; Olson, RE Design and Synthesis of a New Series of 4-Heteroarylamino-1'-azaspiro[oxazole-5,3'-bicyclo[2.2.2]octanes asa7 Nicotinic Receptor Agonists. 1. Development of Pharmacophore and Early Structure-Activity Relationship. J Med Chem59:11171-11181 (2016) [PubMed] Article