

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Histone-lysine N-methyltransferase EZH2 [Y641F] | ||
| Ligand | BDBM172138 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | Enzyme Assay | ||
| Temperature | 298.15±n/a K | ||
| IC50 | 5.53±n/a nM | ||
| Comments | extracted | ||
| Citation |  Kuntz, KW; Chesworth, R; Duncan, KW; Keilhack, H; Warholic, N; Klaus, C; Knutson, SK; Wigle, TJ; Seki, M; Shirotori, S; Kawano, S Aryl- or heteroaryl-substituted benzene compounds US Patent US9090562 Publication Date 7/28/2015 Kuntz, KW; Chesworth, R; Duncan, KW; Keilhack, H; Warholic, N; Klaus, C; Knutson, SK; Wigle, TJ; Seki, M; Shirotori, S; Kawano, S Aryl- or heteroaryl-substituted benzene compounds US Patent US9090562 Publication Date 7/28/2015 | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Histone-lysine N-methyltransferase EZH2 [Y641F] | |||
| Name: | Histone-lysine N-methyltransferase EZH2 [Y641F] | ||
| Synonyms: | EZH2 | EZH2(Y641F) | EZH2_HUMAN | Histone-lysine N-methyltransferase EZH2 (Y641F) | KMT6 | ||
| Type: | n/a | ||
| Mol. Mass.: | 85351.84 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | Q15910[Y641F] | ||
| Residue: | 746 | ||
| Sequence: |
| ||
| BDBM172138 | |||
| n/a | |||
| Name | BDBM172138 | ||
| Synonyms: | EPZ007428 | US10155002, Compound 145 | US11052093, Compound 145 | US9090562, 145 | US9175331, 16 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C30H42N6O2 | ||
| Mol. Mass. | 518.6935 | ||
| SMILES | CCN([C@H]1CC[C@@H](CC1)N(C)C)c1cc(cc(C(=O)NCc2c(C)cc(C)[nH]c2=O)c1C)-c1cnn(C)c1 |r,wU:3.2,wD:6.9,(-2,5.32,;-3.33,4.55,;-3.33,3.01,;-4.67,2.24,;-4.67,.7,;-6,-.07,;-7.34,.7,;-7.34,2.24,;-6,3.01,;-8.67,-.07,;-10,.7,;-8.67,-1.61,;-2,2.24,;-2,.7,;-.67,-.07,;.67,.7,;.67,2.24,;2,3.01,;2,4.55,;3.33,2.24,;4.67,3.01,;6,2.24,;7.34,3.01,;7.34,4.55,;8.67,2.24,;8.67,.7,;10,-.07,;7.34,-.07,;6,.7,;4.67,-.07,;-.67,3.01,;-.67,4.55,;-.67,-1.61,;-1.91,-2.52,;-1.44,-3.98,;.1,-3.98,;.87,-5.32,;.58,-2.52,)| | ||
| Structure | 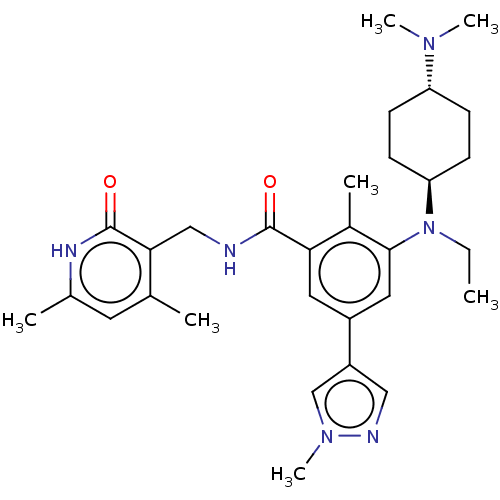
| ||