| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Lysine-specific demethylase 5B |
|---|
| Ligand | BDBM60875 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | Demethylase AlphaScreen Assay |
|---|
| pH | 7.5±n/a |
|---|
| IC50 | 5.5e+2±n/a nM |
|---|
| Comments | extracted |
|---|
| Citation |  Johansson, C; Velupillai, S; Tumber, A; Szykowska, A; Hookway, ES; Nowak, RP; Strain-Damerell, C; Gileadi, C; Philpott, M; Burgess-Brown, N; Wu, N; Kopec, J; Nuzzi, A; Steuber, H; Egner, U; Badock, V; Munro, S; LaThangue, NB; Westaway, S; Brown, J; Athanasou, N; Prinjha, R; Brennan, PE; Oppermann, U Structural analysis of human KDM5B guides histone demethylase inhibitor development. Nat Chem Biol12:539-45 (2016) [PubMed] Article Johansson, C; Velupillai, S; Tumber, A; Szykowska, A; Hookway, ES; Nowak, RP; Strain-Damerell, C; Gileadi, C; Philpott, M; Burgess-Brown, N; Wu, N; Kopec, J; Nuzzi, A; Steuber, H; Egner, U; Badock, V; Munro, S; LaThangue, NB; Westaway, S; Brown, J; Athanasou, N; Prinjha, R; Brennan, PE; Oppermann, U Structural analysis of human KDM5B guides histone demethylase inhibitor development. Nat Chem Biol12:539-45 (2016) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Lysine-specific demethylase 5B |
|---|
| Name: | Lysine-specific demethylase 5B |
|---|
| Synonyms: | CT31 | Cancer/testis antigen 31 | Histone demethylase JARID1B | JARID1B | Jumonji/ARID domain-containing protein 1B | KDM5B | KDM5B_HUMAN | Lysine-specific demethylase 5B (KDM5B) | Lysine-specific demethylase 5B (KDM5Flag-5B-FL) | PLU-1 | PLU1 | RBBP2H1 | RBP2-H1 | Retinoblastoma-binding protein 2 homolog 1 |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 175659.67 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q9UGL1 |
|---|
| Residue: | 1544 |
|---|
| Sequence: | MEAATTLHPGPRPALPLGGPGPLGEFLPPPECPVFEPSWEEFADPFAFIHKIRPIAEQTG
ICKVRPPPDWQPPFACDVDKLHFTPRIQRLNELEAQTRVKLNFLDQIAKYWELQGSTLKI
PHVERKILDLFQLNKLVAEEGGFAVVCKDRKWTKIATKMGFAPGKAVGSHIRGHYERILN
PYNLFLSGDSLRCLQKPNLTTDTKDKEYKPHDIPQRQSVQPSETCPPARRAKRMRAEAMN
IKIEPEETTEARTHNLRRRMGCPTPKCENEKEMKSSIKQEPIERKDYIVENEKEKPKSRS
KKATNAVDLYVCLLCGSGNDEDRLLLCDGCDDSYHTFCLIPPLHDVPKGDWRCPKCLAQE
CSKPQEAFGFEQAARDYTLRTFGEMADAFKSDYFNMPVHMVPTELVEKEFWRLVSTIEED
VTVEYGADIASKEFGSGFPVRDGKIKLSPEEEEYLDSGWNLNNMPVMEQSVLAHITADIC
GMKLPWLYVGMCFSSFCWHIEDHWSYSINYLHWGEPKTWYGVPGYAAEQLENVMKKLAPE
LFVSQPDLLHQLVTIMNPNTLMTHEVPVYRTNQCAGEFVITFPRAYHSGFNQGFNFAEAV
NFCTVDWLPLGRQCVEHYRLLHRYCVFSHDEMICKMASKADVLDVVVASTVQKDMAIMIE
DEKALRETVRKLGVIDSERMDFELLPDDERQCVKCKTTCFMSAISCSCKPGLLVCLHHVK
ELCSCPPYKYKLRYRYTLDDLYPMMNALKLRAESYNEWALNVNEALEAKINKKKSLVSFK
ALIEESEMKKFPDNDLLRHLRLVTQDAEKCASVAQQLLNGKRQTRYRSGGGKSQNQLTVN
ELRQFVTQLYALPCVLSQTPLLKDLLNRVEDFQQHSQKLLSEETPSAAELQDLLDVSFEF
DVELPQLAEMRIRLEQARWLEEVQQACLDPSSLTLDDMRRLIDLGVGLAPYSAVEKAMAR
LQELLTVSEHWDDKAKSLLKARPRHSLNSLATAVKEIEEIPAYLPNGAALKDSVQRARDW
LQDVEGLQAGGRVPVLDTLIELVTRGRSIPVHLNSLPRLETLVAEVQAWKECAVNTFLTE
NSPYSLLEVLCPRCDIGLLGLKRKQRKLKEPLPNGKKKSTKLESLSDLERALTESKETAS
AMATLGEARLREMEALQSLRLANEGKLLSPLQDVDIKICLCQKAPAAPMIQCELCRDAFH
TSCVAVPSISQGLRIWLCPHCRRSEKPPLEKILPLLASLQRIRVRLPEGDALRYMIERTV
NWQHRAQQLLSSGNLKFVQDRVGSGLLYSRWQASAGQVSDTNKVSQPPGTTSFSLPDDWD
NRTSYLHSPFSTGRSCIPLHGVSPEVNELLMEAQLLQVSLPEIQELYQTLLAKPSPAQQT
DRSSPVRPSSEKNDCCRGKRDGINSLERKLKRRLEREGLSSERWERVKKMRTPKKKKIKL
SHPKDMNNFKLERERSYELVRSAETHSLPSDTSYSEQEDSEDEDAICPAVSCLQPEGDEV
DWVQCDGSCNQWFHQVCVGVSPEMAEKEDYICVRCTVKDAPSRK
|
|
|
|---|
| BDBM60875 |
|---|
| n/a |
|---|
| Name | BDBM60875 |
|---|
| Synonyms: | 3-((6-(4,5-Dihydro-1H-benzo[d]azepin-3(2H)-yl)-2-(pyridin-2-yl)pyrimidin-4-yl)amino)propanoic acid | 3-{[2-(pyridin-2-yl)-6-(2,3,4,5-tetrahydro-1H-3-benzazepin-3-yl)pyrimidin-4-yl]amino}propanoic acid | GSK J1 | GSK-J1 | GSKJ1 |
|---|
| Type | n/a |
|---|
| Emp. Form. | C22H23N5O2 |
|---|
| Mol. Mass. | 389.4503 |
|---|
| SMILES | OC(=O)CCNc1cc(nc(n1)-c1ccccn1)N1CCc2ccccc2CC1 |
|---|
| Structure | 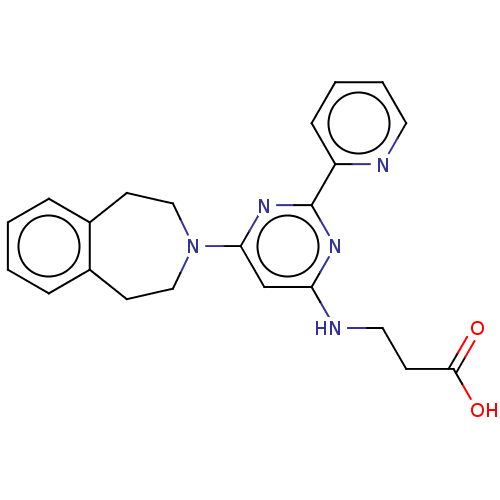
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Johansson, C; Velupillai, S; Tumber, A; Szykowska, A; Hookway, ES; Nowak, RP; Strain-Damerell, C; Gileadi, C; Philpott, M; Burgess-Brown, N; Wu, N; Kopec, J; Nuzzi, A; Steuber, H; Egner, U; Badock, V; Munro, S; LaThangue, NB; Westaway, S; Brown, J; Athanasou, N; Prinjha, R; Brennan, PE; Oppermann, U Structural analysis of human KDM5B guides histone demethylase inhibitor development. Nat Chem Biol12:539-45 (2016) [PubMed] Article
Johansson, C; Velupillai, S; Tumber, A; Szykowska, A; Hookway, ES; Nowak, RP; Strain-Damerell, C; Gileadi, C; Philpott, M; Burgess-Brown, N; Wu, N; Kopec, J; Nuzzi, A; Steuber, H; Egner, U; Badock, V; Munro, S; LaThangue, NB; Westaway, S; Brown, J; Athanasou, N; Prinjha, R; Brennan, PE; Oppermann, U Structural analysis of human KDM5B guides histone demethylase inhibitor development. Nat Chem Biol12:539-45 (2016) [PubMed] Article