

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Neuronal acetylcholine receptor subunit alpha-4/beta-2 (Rattus norvegicus (Rat)) | BDBM50143314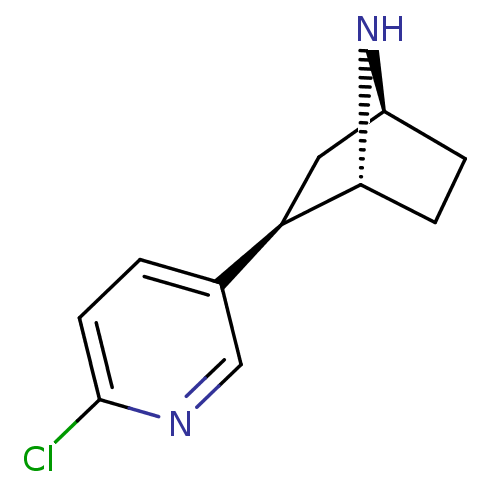 ((+)-Epibatidine | (-)-epibatidine | (1R,2R,4S)-2-(...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL DrugBank MMDB PC cid PC sid PDB UniChem Patents Similars | Article PubMed | 0 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Universit£ di Sassari Curated by ChEMBL | Assay Description Displacement of [3H]epibatidine from alpha4beta2 nAChR in rat cerebral cortex membrane | Eur J Med Chem 180: 51-61 (2019) Article DOI: 10.1016/j.ejmech.2019.06.079 BindingDB Entry DOI: 10.7270/Q24Q7ZBT | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Gag-Pol polyprotein [489-587] (Human immunodeficiency virus type 1) | BDBM4690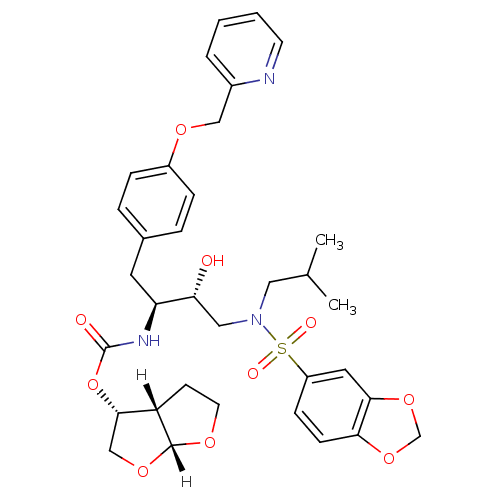 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.00000600 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
GlaxoSmithKline Curated by ChEMBL | Assay Description Displacement of [3H]GW0385 from HIV1 protease | Bioorg Med Chem Lett 16: 1788-94 (2006) Article DOI: 10.1016/j.bmcl.2006.01.035 BindingDB Entry DOI: 10.7270/Q2WS8V1F | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Gag-Pol polyprotein [489-587] (Human immunodeficiency virus type 1) | BDBM4685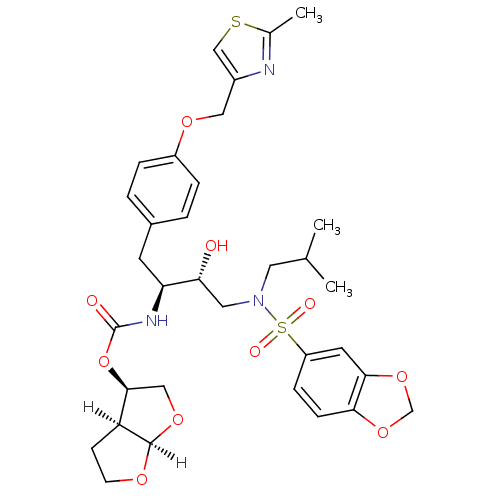 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 0.0000150 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
GlaxoSmithKline Curated by ChEMBL | Assay Description Displacement of [3H]GW0385 from HIV1 protease | Bioorg Med Chem Lett 16: 1788-94 (2006) Article DOI: 10.1016/j.bmcl.2006.01.035 BindingDB Entry DOI: 10.7270/Q2WS8V1F | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Sigma non-opioid intracellular receptor 1 (Homo sapiens (Human)) | BDBM50398057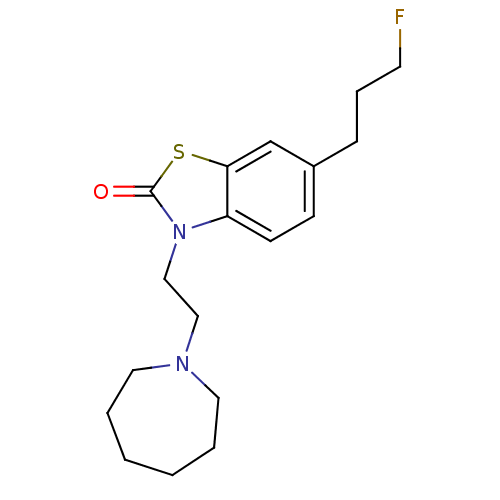 (CHEMBL2181924) | PDB UniProtKB/SwissProt antibodypedia GoogleScholar AffyNet | Purchase CHEMBL PC cid PC sid UniChem Similars | 0.00250 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a | |
TBA | Assay Description In vitro inhibitory activity against Angiotensin I converting enzyme | Citation and Details | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Prothrombin (Homo sapiens (Human)) | BDBM50133524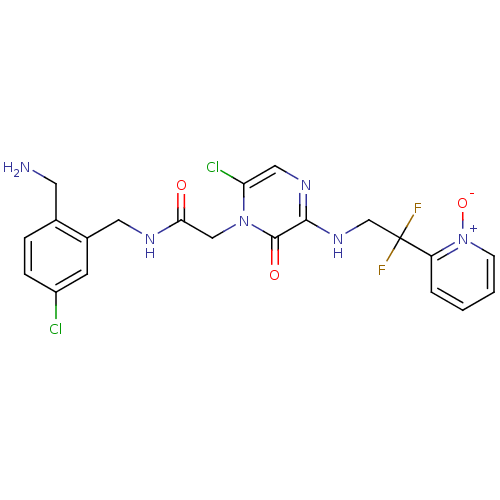 (CHEMBL419773 | N-(2-Aminomethyl-5-chloro-benzyl)-2...) | PDB UniProtKB/SwissProt antibodypedia GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | 0.00340 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Merck Research Laboratories Curated by ChEMBL | Assay Description Inhibitory constant evaluated against thrombin (Factor IIa) | Bioorg Med Chem Lett 13: 3477-82 (2003) BindingDB Entry DOI: 10.7270/Q29P3119 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17129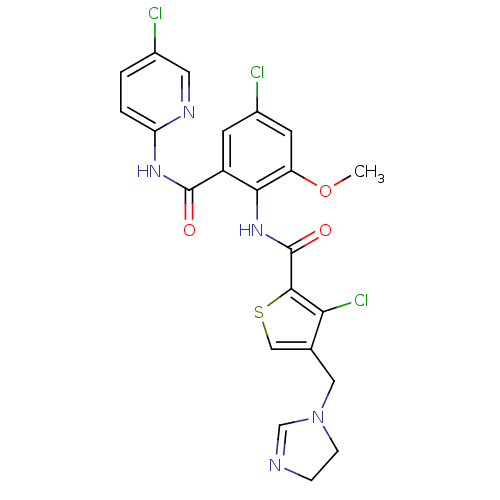 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.00400 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17122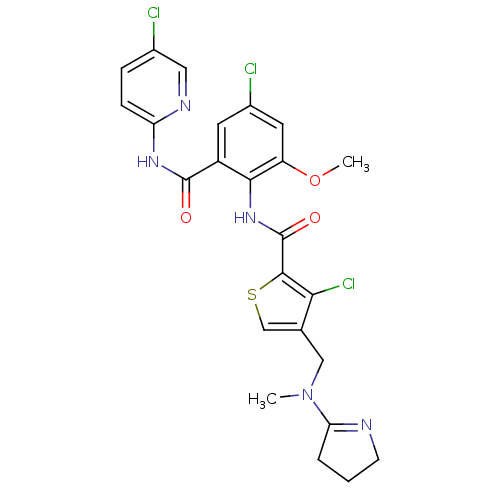 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.00400 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17127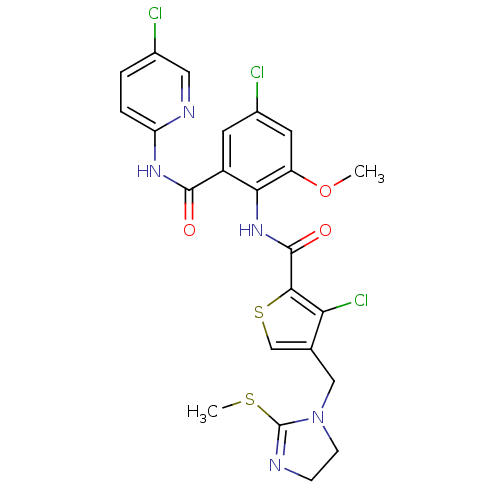 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.00400 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A] (Human immunodeficiency virus type 1) | BDBM9182 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | <0.00500 | n/a | 3 | n/a | n/a | n/a | n/a | n/a | n/a |
GlaxoSmithKline | Assay Description The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f... | Bioorg Med Chem Lett 15: 3560-4 (2005) Article DOI: 10.1016/j.bmcl.2005.05.101 BindingDB Entry DOI: 10.7270/Q2F769SC | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A] (Human immunodeficiency virus type 1) | BDBM9180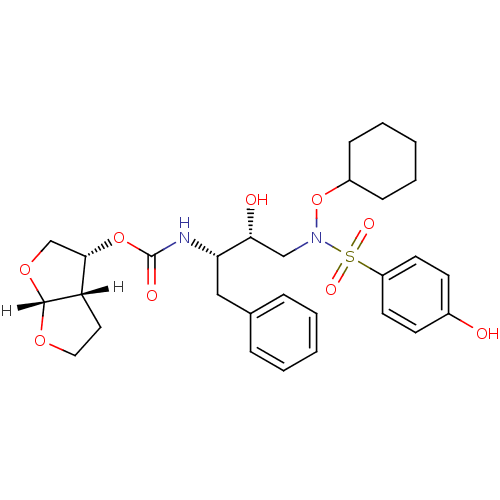 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | <0.00500 | n/a | 5 | n/a | n/a | n/a | n/a | n/a | n/a |
GlaxoSmithKline | Assay Description The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f... | Bioorg Med Chem Lett 15: 3560-4 (2005) Article DOI: 10.1016/j.bmcl.2005.05.101 BindingDB Entry DOI: 10.7270/Q2F769SC | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A] (Human immunodeficiency virus type 1) | BDBM9171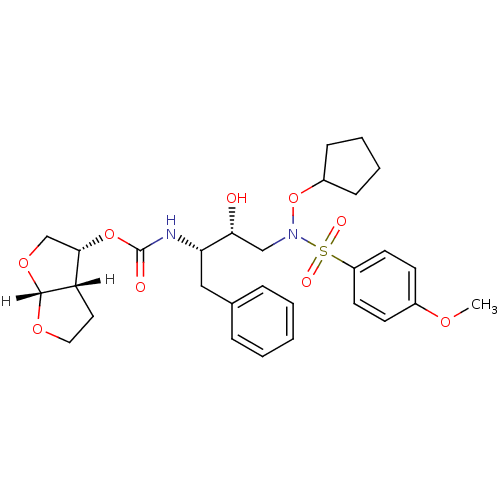 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | <0.00500 | <-64.5 | 6 | n/a | n/a | n/a | n/a | 6.8 | 25 |
GlaxoSmithKline | Assay Description The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f... | Bioorg Med Chem Lett 15: 3560-4 (2005) Article DOI: 10.1016/j.bmcl.2005.05.101 BindingDB Entry DOI: 10.7270/Q2F769SC | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A] (Human immunodeficiency virus type 1) | BDBM9175 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | <0.00500 | n/a | 7 | n/a | n/a | n/a | n/a | n/a | n/a |
GlaxoSmithKline | Assay Description The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f... | Bioorg Med Chem Lett 15: 3560-4 (2005) Article DOI: 10.1016/j.bmcl.2005.05.101 BindingDB Entry DOI: 10.7270/Q2F769SC | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A] (Human immunodeficiency virus type 1) | BDBM9176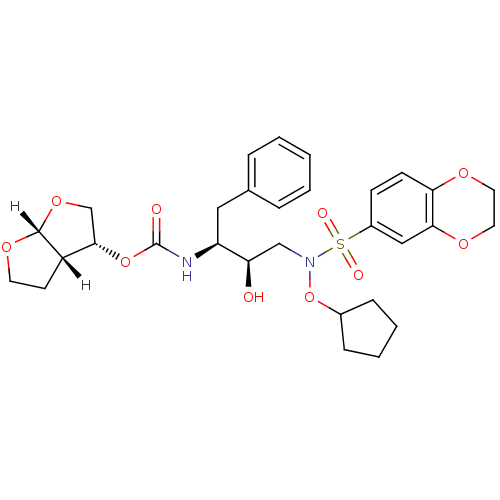 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | <0.00500 | n/a | 7 | n/a | n/a | n/a | n/a | n/a | n/a |
GlaxoSmithKline | Assay Description The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f... | Bioorg Med Chem Lett 15: 3560-4 (2005) Article DOI: 10.1016/j.bmcl.2005.05.101 BindingDB Entry DOI: 10.7270/Q2F769SC | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A] (Human immunodeficiency virus type 1) | BDBM9173 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | <0.00500 | n/a | 9 | n/a | n/a | n/a | n/a | n/a | n/a |
GlaxoSmithKline | Assay Description The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f... | Bioorg Med Chem Lett 15: 3560-4 (2005) Article DOI: 10.1016/j.bmcl.2005.05.101 BindingDB Entry DOI: 10.7270/Q2F769SC | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A] (Human immunodeficiency virus type 1) | BDBM9178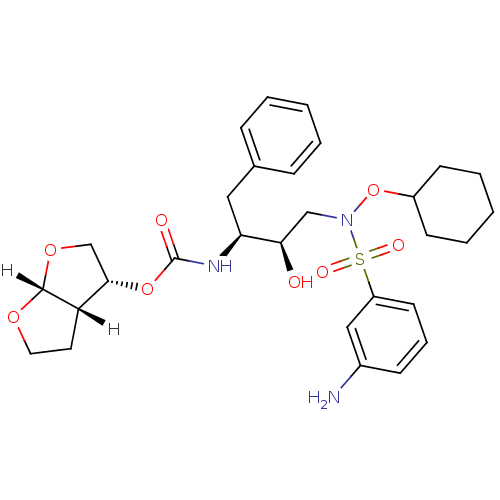 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.00500 | n/a | 11 | n/a | n/a | n/a | n/a | n/a | n/a |
GlaxoSmithKline | Assay Description The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f... | Bioorg Med Chem Lett 15: 3560-4 (2005) Article DOI: 10.1016/j.bmcl.2005.05.101 BindingDB Entry DOI: 10.7270/Q2F769SC | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A] (Human immunodeficiency virus type 1) | BDBM9174 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | <0.00500 | n/a | 12 | n/a | n/a | n/a | n/a | n/a | n/a |
GlaxoSmithKline | Assay Description The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f... | Bioorg Med Chem Lett 15: 3560-4 (2005) Article DOI: 10.1016/j.bmcl.2005.05.101 BindingDB Entry DOI: 10.7270/Q2F769SC | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A] (Human immunodeficiency virus type 1) | BDBM9181 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | <0.00500 | n/a | 13 | n/a | n/a | n/a | n/a | n/a | n/a |
GlaxoSmithKline | Assay Description The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f... | Bioorg Med Chem Lett 15: 3560-4 (2005) Article DOI: 10.1016/j.bmcl.2005.05.101 BindingDB Entry DOI: 10.7270/Q2F769SC | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A] (Human immunodeficiency virus type 1) | BDBM9172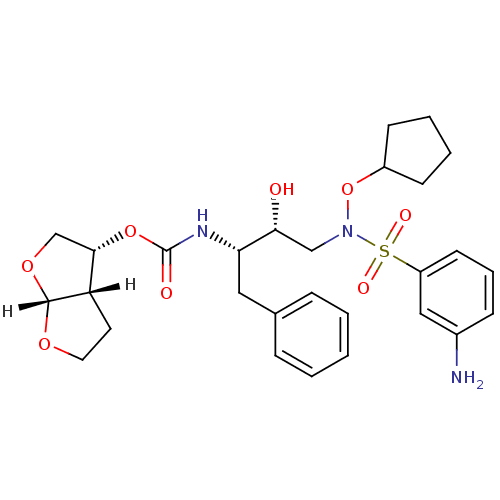 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | <0.00500 | n/a | 17 | n/a | n/a | n/a | n/a | n/a | n/a |
GlaxoSmithKline | Assay Description The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f... | Bioorg Med Chem Lett 15: 3560-4 (2005) Article DOI: 10.1016/j.bmcl.2005.05.101 BindingDB Entry DOI: 10.7270/Q2F769SC | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A] (Human immunodeficiency virus type 1) | BDBM9177 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | <0.00500 | n/a | 20 | n/a | n/a | n/a | n/a | n/a | n/a |
GlaxoSmithKline | Assay Description The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f... | Bioorg Med Chem Lett 15: 3560-4 (2005) Article DOI: 10.1016/j.bmcl.2005.05.101 BindingDB Entry DOI: 10.7270/Q2F769SC | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A] (Human immunodeficiency virus type 1) | BDBM9170 ((2S)-N-[(2S,3R)-4-[(cyclopentyloxy)(4-methoxybenze...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | <0.00500 | <-64.5 | 24 | n/a | n/a | n/a | n/a | 6.8 | 25 |
GlaxoSmithKline | Assay Description The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f... | Bioorg Med Chem Lett 15: 3560-4 (2005) Article DOI: 10.1016/j.bmcl.2005.05.101 BindingDB Entry DOI: 10.7270/Q2F769SC | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17136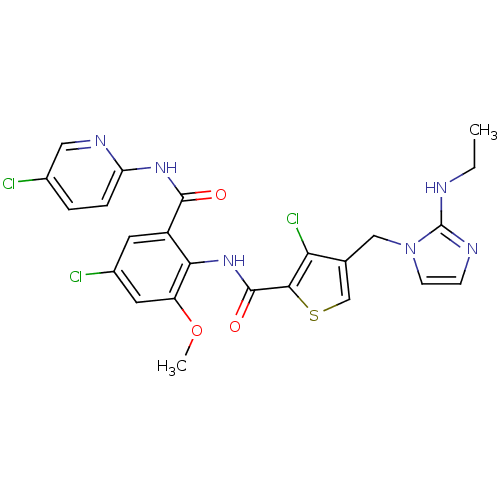 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.00500 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17135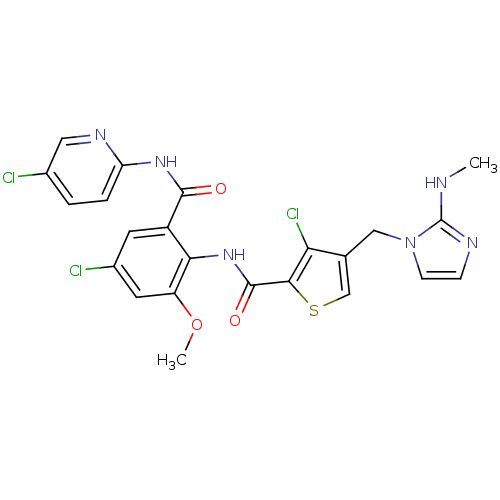 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | MMDB PC cid PC sid PDB UniChem Patents Similars | MMDB PDB Article PubMed | 0.00500 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17134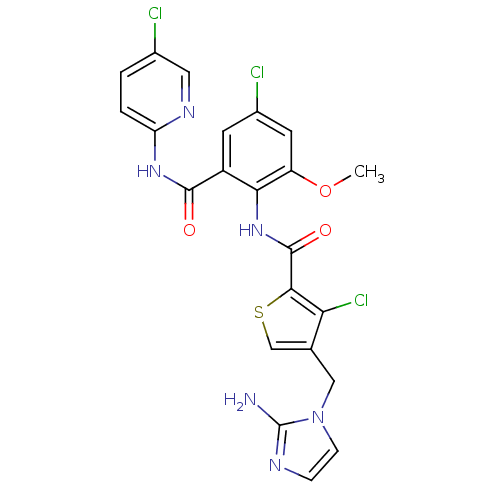 (4-[(2-amino-1H-imidazol-1-yl)methyl]-3-chloro-N-{4...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.00600 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Melatonin receptor type 1A/1B (Homo sapiens (Human)) | BDBM50118462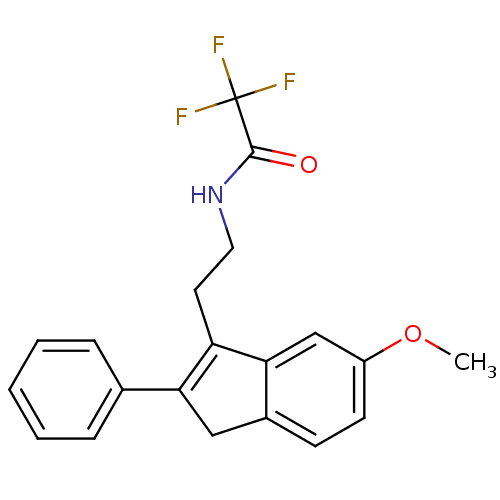 (2,2,2-Trifluoro-N-[2-(6-methoxy-2-phenyl-3H-inden-...) | PDB KEGG UniProtKB/SwissProt DrugBank antibodypedia antibodypedia GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents Similars | PubMed | 0.00602 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Takeda Chemical Industries, Ltd. Curated by ChEMBL | Assay Description Ability to inhibit 2-[125I]iodomelatonin specific binding to human melatonin receptor type 1A (MT1) expressed in CHO cells. | J Med Chem 45: 4212-21 (2002) BindingDB Entry DOI: 10.7270/Q2J102HG | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Bone morphogenetic protein 1 (Homo sapiens (Human)) | BDBM50458766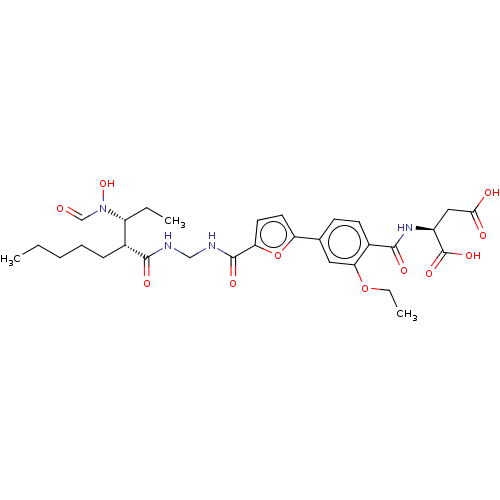 (CHEMBL4212386) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD antibodypedia GoogleScholar AffyNet | KEGG PC cid PC sid PDB UniChem | PDB Article PubMed | 0.00680 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
GlaxoSmithKline Pharmaceuticals Curated by ChEMBL | Assay Description Binding affinity to BMP1 (unknown origin) using ((5-FAM)-ELIDQYDVQRDDSSDGSLED-K(5,6 TAMRA)-CONH2 as substrate preincubated for 3 hrs followed by subs... | ACS Med Chem Lett 9: 736-740 (2018) Article DOI: 10.1021/acsmedchemlett.8b00173 BindingDB Entry DOI: 10.7270/Q2X35127 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17111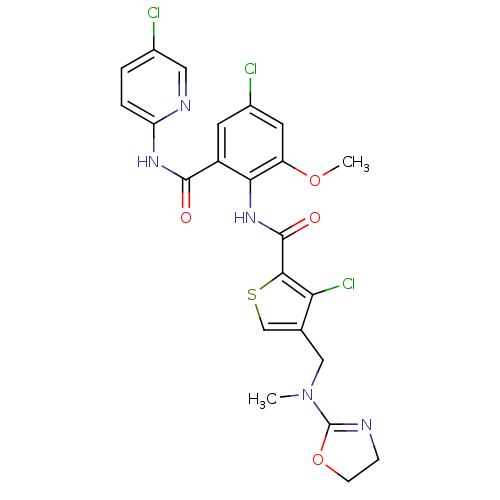 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid PDB UniChem Similars | PDB Article PubMed | 0.00700 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17137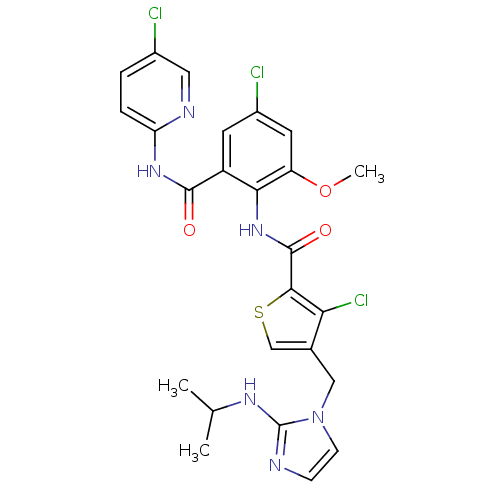 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 0.00800 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Endothelin receptor type B (Sus scrofa) | BDBM50000558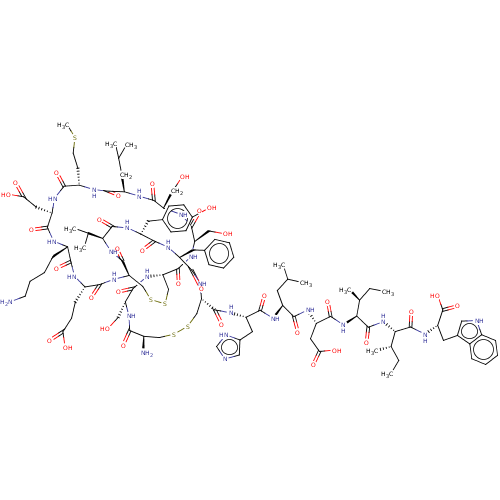 (CHEMBL437472 | ET-1 | Endothelin -1 | Endothelin 1...) | PDB Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article | 0.00800 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description Ability to inhibit [125I]ET-3 binding to the Endothelin B receptor from porcine lung membranes was evaluated | Bioorg Med Chem Lett 6: 2323-2328 (1996) Article DOI: 10.1016/0960-894X(96)00421-0 BindingDB Entry DOI: 10.7270/Q21J9B8K | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| 5-hydroxytryptamine receptor 2A (Rattus norvegicus (rat)) | BDBM84928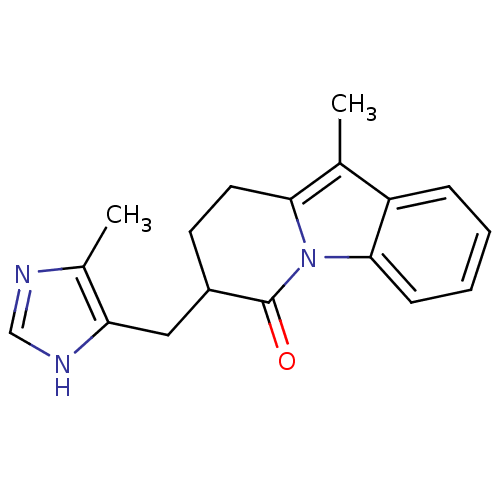 (CAS_125368 | FK 1052 | NSC_125368) | PDB Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | Purchase MCE PC cid PC sid UniChem Patents | PubMed | 0.0100 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Fujisawa Pharmaceutical Co., Ltd. Curated by PDSP Ki Database | J Pharmacol Exp Ther 265: 752-8 (1993) BindingDB Entry DOI: 10.7270/Q2CV4G73 | ||||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Neuronal acetylcholine receptor subunit alpha-4/beta-2 (Rattus norvegicus (Rat)) | BDBM50515333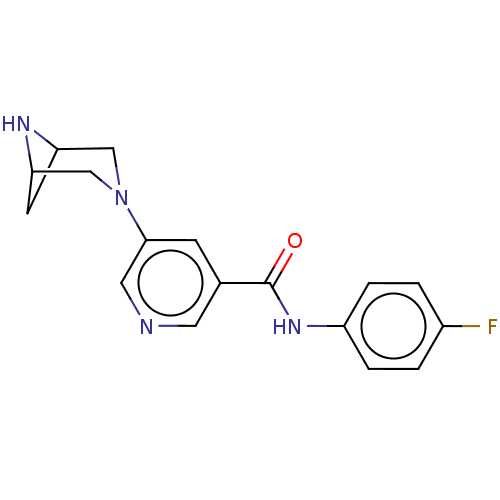 (CHEMBL4460815) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 0.0100 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Universit£ di Sassari Curated by ChEMBL | Assay Description Displacement of [3H]epibatidine from alpha4beta2 nAChR in rat cerebral cortex membrane | Eur J Med Chem 180: 51-61 (2019) Article DOI: 10.1016/j.ejmech.2019.06.079 BindingDB Entry DOI: 10.7270/Q24Q7ZBT | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| D(2) dopamine receptor (Rattus norvegicus (rat)) | BDBM84928 (CAS_125368 | FK 1052 | NSC_125368) | PDB Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | Purchase MCE PC cid PC sid UniChem Patents | PubMed | 0.0100 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Fujisawa Pharmaceutical Co., Ltd. Curated by PDSP Ki Database | J Pharmacol Exp Ther 265: 752-8 (1993) BindingDB Entry DOI: 10.7270/Q2CV4G73 | ||||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A] (Human immunodeficiency virus type 1) | BDBM518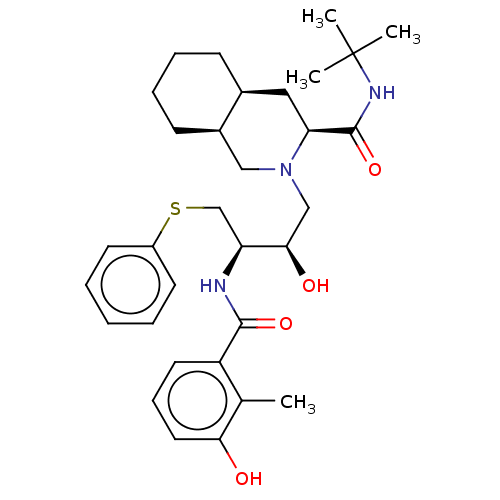 ((3S,4aS,8aS)-N-tert-butyl-2-[(2R,3R)-2-hydroxy-3-[...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL MCE MMDB PC cid PC sid PDB UniChem Patents Similars | MMDB PDB Article PubMed | 0.0100 | -62.8 | n/a | n/a | n/a | n/a | n/a | 6.4 | 25 |
GlaxoSmithKline | Assay Description The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f... | Bioorg Med Chem Lett 10: 1159-62 (2000) Article DOI: 10.1016/s0960-894x(00)00163-3 BindingDB Entry DOI: 10.7270/Q2X63K5G | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17123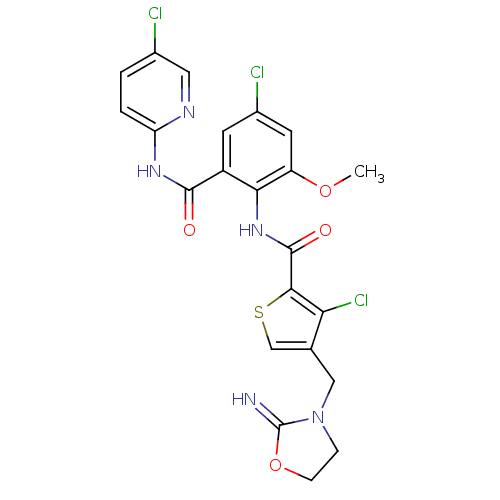 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.0100 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17133 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.0100 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17124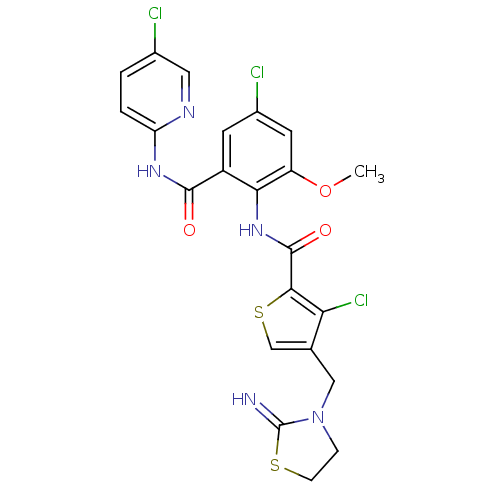 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.0100 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17125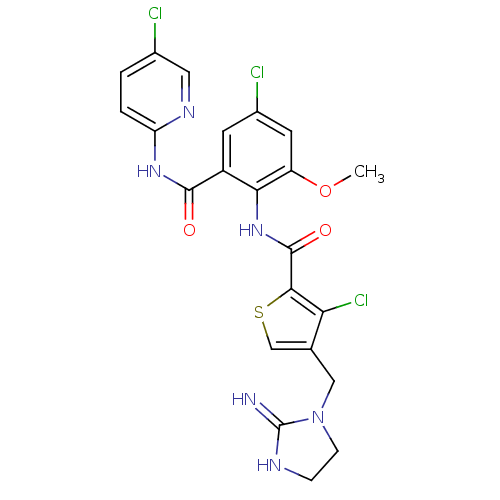 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.0100 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17128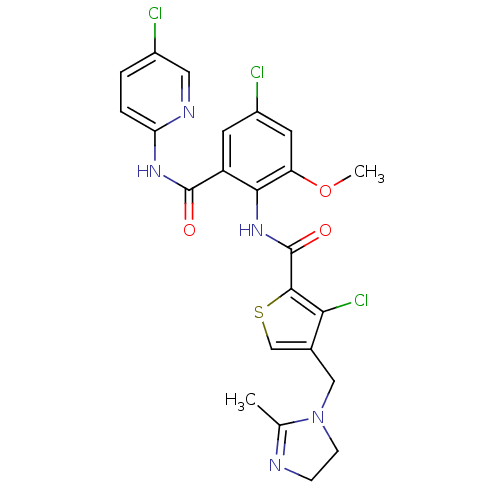 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.0100 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17130 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.0100 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17131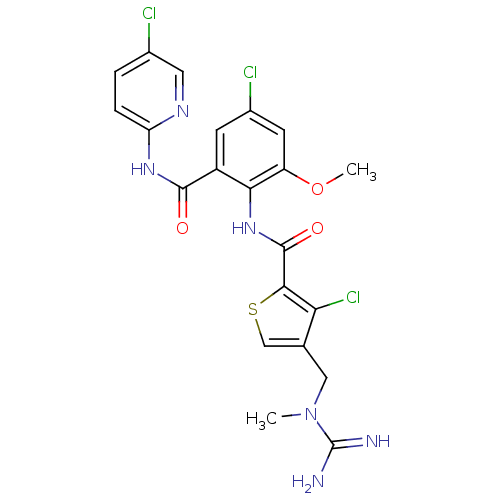 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.0100 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Sigma non-opioid intracellular receptor 1 (RAT) | BDBM50170654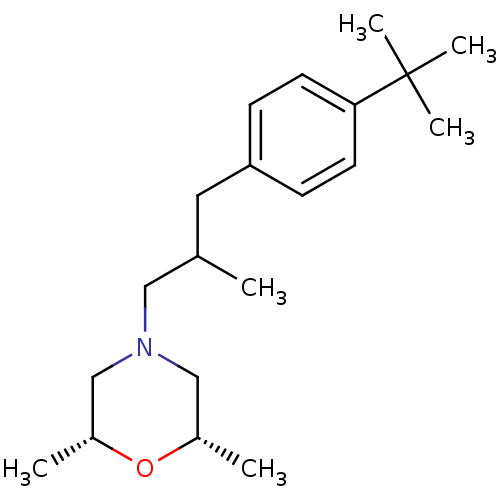 ((+-)-cis-4-(3-(4-tert-butylphenyl)-2-methylpropyl)...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | Purchase CHEMBL MCE KEGG PC cid PC sid UniChem Patents | Article PubMed | 0.0110 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description Displacement of [3H]-pentazocine from sigma 1 opioid receptor in rat liver homogenate by liquid scintillation counting | ACS Med Chem Lett 2: 834-839 (2011) Article DOI: 10.1021/ml2001505 BindingDB Entry DOI: 10.7270/Q2RR208C | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17118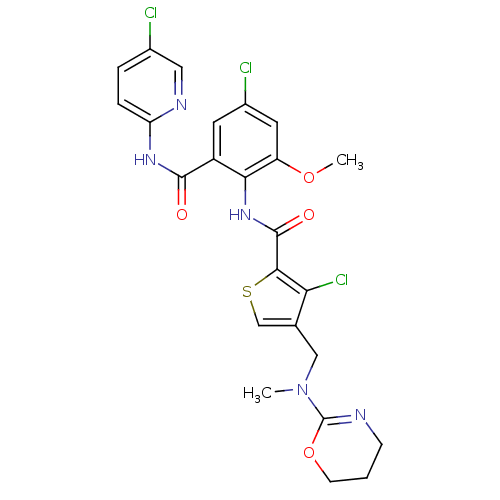 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 0.0120 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17112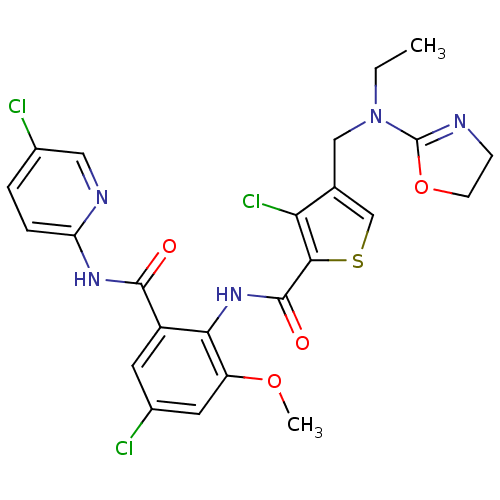 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.0120 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Melatonin receptor type 1A/1B (Homo sapiens (Human)) | BDBM50118435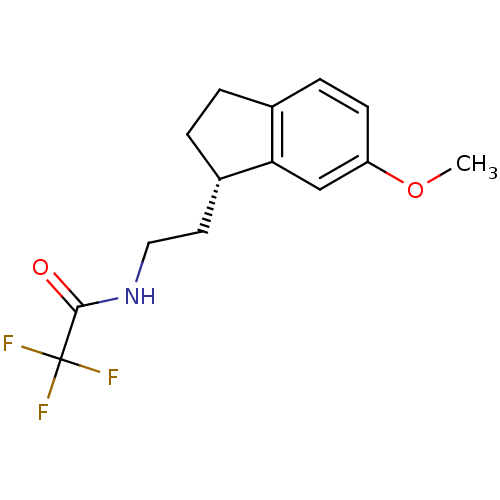 (2,2,2-Trifluoro-N-[2-(6-methoxy-indan-1-yl)-ethyl]...) | PDB KEGG UniProtKB/SwissProt DrugBank antibodypedia antibodypedia GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents Similars | PubMed | 0.0123 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Takeda Chemical Industries, Ltd. Curated by ChEMBL | Assay Description Ability to inhibit 2-[125I]iodomelatonin specific binding to human melatonin receptor type 1A (MT1) expressed in CHO cells. | J Med Chem 45: 4212-21 (2002) BindingDB Entry DOI: 10.7270/Q2J102HG | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Melatonin receptor type 1A (Homo sapiens (Human)) | BDBM50118470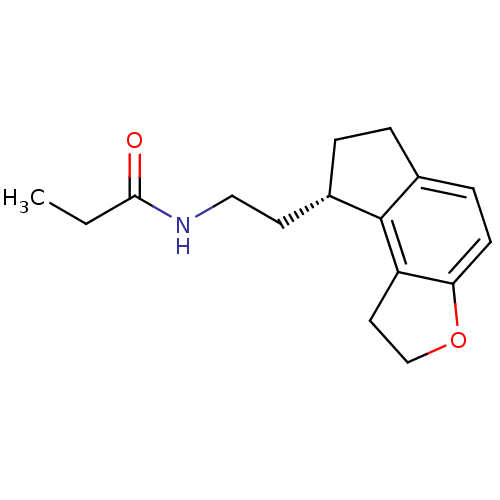 (CHEMBL1218 | N-[2-(1,6,7,8-Tetrahydro-2H-3-oxa-as-...) | PDB Reactome pathway KEGG UniProtKB/SwissProt DrugBank antibodypedia GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE PC cid PC sid PDB UniChem Patents Similars | DrugBank PDB PubMed | 0.0138 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Takeda Chemical Industries, Ltd. Curated by ChEMBL | Assay Description Binding affinity against human Melatonin receptor type 1A (MT1) | J Med Chem 45: 4222-39 (2002) BindingDB Entry DOI: 10.7270/Q2D799S6 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Melatonin receptor type 1B (Homo sapiens (Human)) | BDBM50240440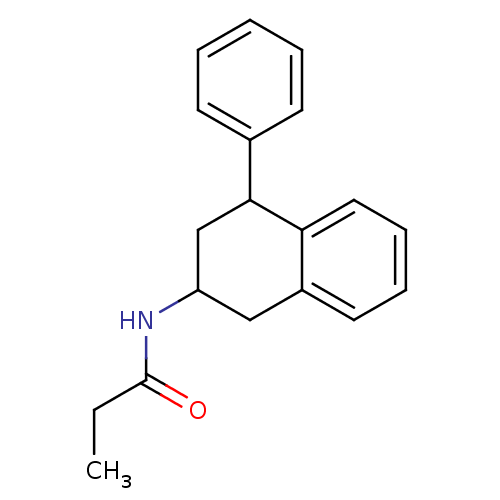 (4-P-PDOT | CHEMBL285718 | N-(4-Phenyl-1,2,3,4-tetr...) | PDB Reactome pathway KEGG UniProtKB/SwissProt DrugBank antibodypedia GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid UniChem Patents Similars | Article PubMed | 0.0158 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Universit£ degli Studi di Parma Curated by ChEMBL | Assay Description Inhibition of 2-[125I]iodomelatonin binding to human melatonin receptor MT2 expressed in NIH3T3 rat fibroblast cells | J Med Chem 48: 4049-60 (2005) Article DOI: 10.1021/jm048956y BindingDB Entry DOI: 10.7270/Q2319ZND | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Melatonin receptor type 1A/1B (Homo sapiens (Human)) | BDBM50034110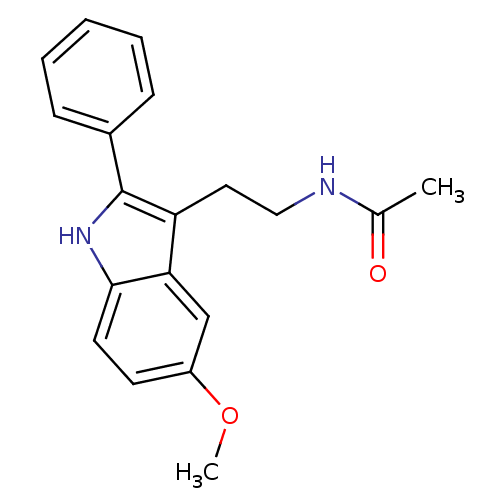 (CHEMBL15060 | Melatonin,2-Phenyl | N-[2-(5-Methoxy...) | PDB KEGG UniProtKB/SwissProt DrugBank antibodypedia antibodypedia GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 0.0170 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Universit£ degli Studi di Urbino Curated by ChEMBL | Assay Description Binding affinity against melatonin receptor in the quail optica tecta using 2-[125] iodomelatonin as labelled ligand | J Med Chem 40: 1990-2002 (1997) Article DOI: 10.1021/jm960651z BindingDB Entry DOI: 10.7270/Q2HD7ZCK | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Dimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A] (Human immunodeficiency virus type 1) | BDBM9179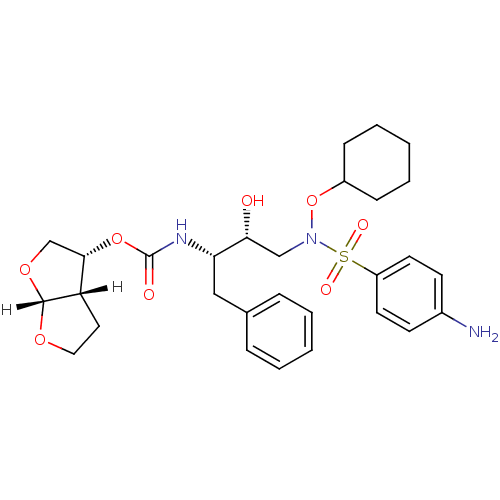 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.0180 | n/a | 8 | n/a | n/a | n/a | n/a | n/a | n/a |
GlaxoSmithKline | Assay Description The Ki values were determined using fluorogenic substrate, 2-(aminobenzoyl)-Thr-Ile-Nle-Phe(p-NO2)-Gln-ArgNH2. A standard curve relating changes in f... | Bioorg Med Chem Lett 15: 3560-4 (2005) Article DOI: 10.1016/j.bmcl.2005.05.101 BindingDB Entry DOI: 10.7270/Q2F769SC | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17104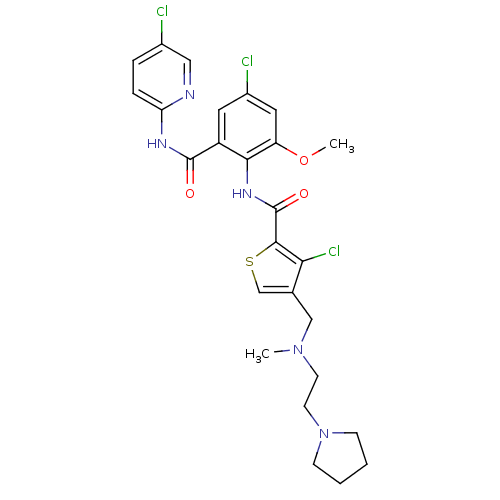 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.0190 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17138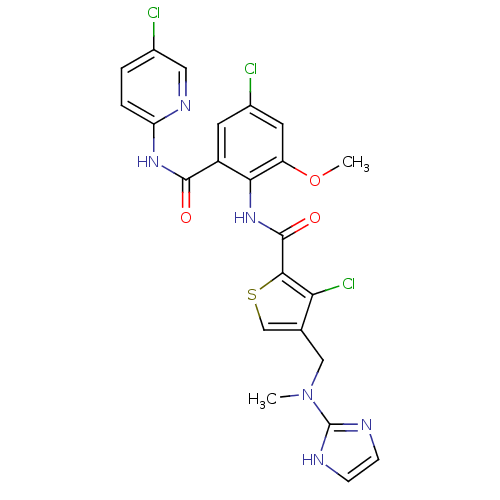 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 0.0200 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Tyrosine-protein kinase ABL1 (Homo sapiens (Human)) | BDBM50378812 (CHEMBL1221411) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 0.0200 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Iowa State University Curated by ChEMBL | Assay Description Inhibition of recombinant c-Abl after 30 mins | Bioorg Med Chem 18: 6316-21 (2010) Article DOI: 10.1016/j.bmc.2010.07.021 BindingDB Entry DOI: 10.7270/Q2CZ384X | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Displayed 1 to 50 (of 98480 total ) | Next | Last >> |