

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dimer of Gag-Pol polyprotein [489-587] (Human immunodeficiency virus type 1) | BDBM200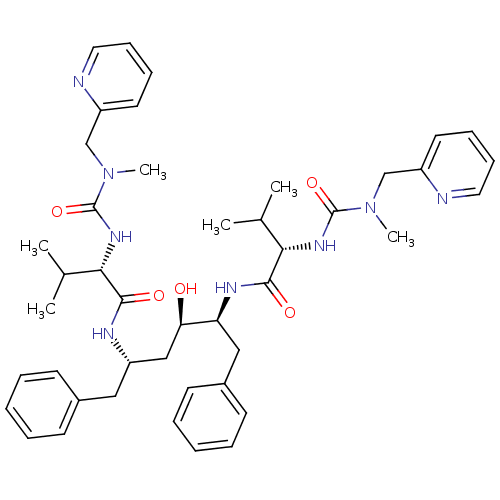 ((2S)-N-[(2S,3R,5S)-3-hydroxy-5-[(2S)-3-methyl-2-{[...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid PDB UniChem Patents Similars | PDB Article | 0.00400 | -66.1 | n/a | n/a | n/a | n/a | n/a | 4.7 | 30 |
NCI-FCRDC | Assay Description HIV-1 protease activity was measured by a continuous fluorometric assay using the internally quenched fluorogenic substrate DABCYL-GABA-Ser-Gln-Tyr-P... | J Am Chem Soc 116: 847-55 (1994) Article DOI: 10.1021/ja00082a004 BindingDB Entry DOI: 10.7270/Q2KK98Z1 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Dimer of Gag-Pol polyprotein [489-587] (Human immunodeficiency virus type 1) | BDBM198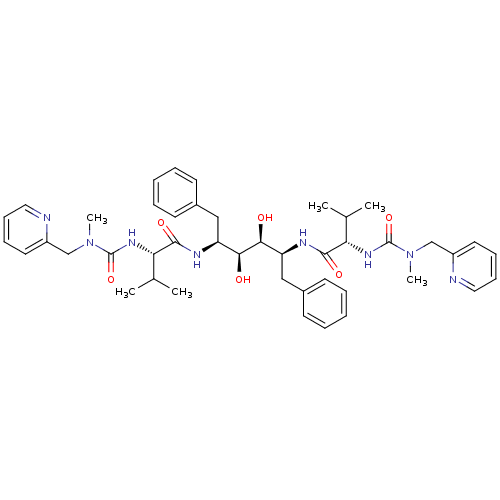 ((2S)-N-[(2S,3S,4S,5S)-3,4-dihydroxy-5-[(2S)-3-meth...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | MMDB PC cid PC sid PDB UniChem Similars | PDB Article | 0.0110 | -63.6 | n/a | n/a | n/a | n/a | n/a | 4.7 | 30 |
NCI-FCRDC | Assay Description HIV-1 protease activity was measured by a continuous fluorometric assay using the internally quenched fluorogenic substrate DABCYL-GABA-Ser-Gln-Tyr-P... | J Am Chem Soc 116: 847-55 (1994) Article DOI: 10.1021/ja00082a004 BindingDB Entry DOI: 10.7270/Q2KK98Z1 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Dimer of Gag-Pol polyprotein [489-587] (Human immunodeficiency virus type 1) | BDBM199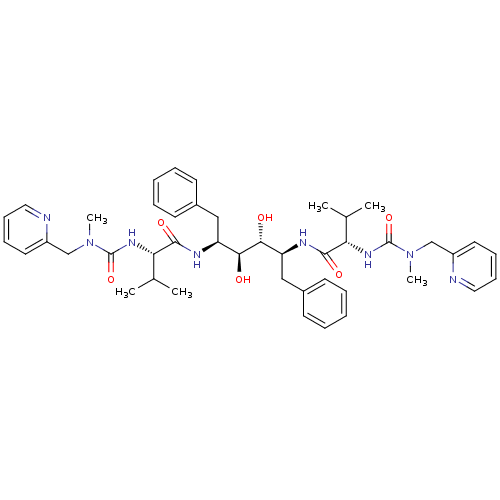 ((2S)-N-[(2S,3R,4S,5S)-3,4-dihydroxy-5-[(2S)-3-meth...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | KEGG MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article | 0.0120 | -63.4 | n/a | n/a | n/a | n/a | n/a | 4.7 | 30 |
NCI-FCRDC | Assay Description HIV-1 protease activity was measured by a continuous fluorometric assay using the internally quenched fluorogenic substrate DABCYL-GABA-Ser-Gln-Tyr-P... | J Am Chem Soc 116: 847-55 (1994) Article DOI: 10.1021/ja00082a004 BindingDB Entry DOI: 10.7270/Q2KK98Z1 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Dimer of Gag-Pol polyprotein [489-587] (Human immunodeficiency virus type 1) | BDBM197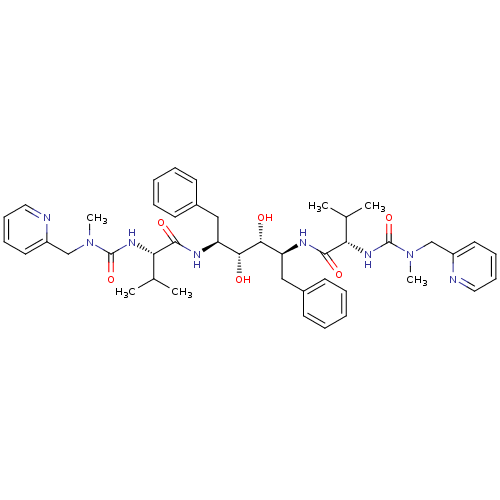 ((2S)-N-[(2S,3R,4R,5S)-3,4-dihydroxy-5-[(2S)-3-meth...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article | 0.112 | -57.7 | n/a | n/a | n/a | n/a | n/a | 4.7 | 30 |
NCI-FCRDC | Assay Description HIV-1 protease activity was measured by a continuous fluorometric assay using the internally quenched fluorogenic substrate DABCYL-GABA-Ser-Gln-Tyr-P... | J Am Chem Soc 116: 847-55 (1994) Article DOI: 10.1021/ja00082a004 BindingDB Entry DOI: 10.7270/Q2KK98Z1 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Muscarinic acetylcholine receptor M2 (RAT) | BDBM50453895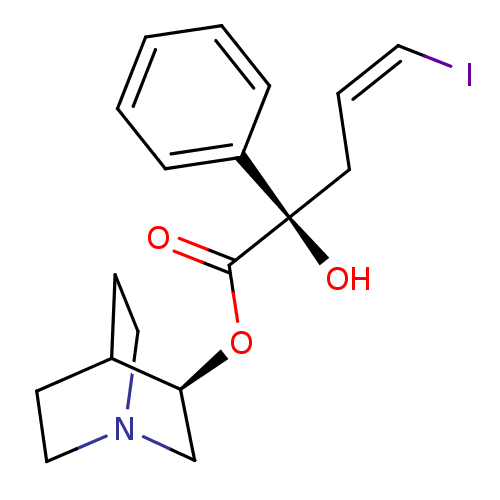 (CHEMBL2111842) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | n/a | n/a | n/a | 0.430 | n/a | n/a | n/a | n/a | n/a |
Oak Ridge National Laboratory (ORNL) Curated by ChEMBL | Assay Description In vitro binding affinity towards Muscarinic acetylcholine receptor M2 using [3H]-QNB as radioligand from rat heart tissue | J Med Chem 38: 3908-17 (1995) BindingDB Entry DOI: 10.7270/Q24M9567 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50450592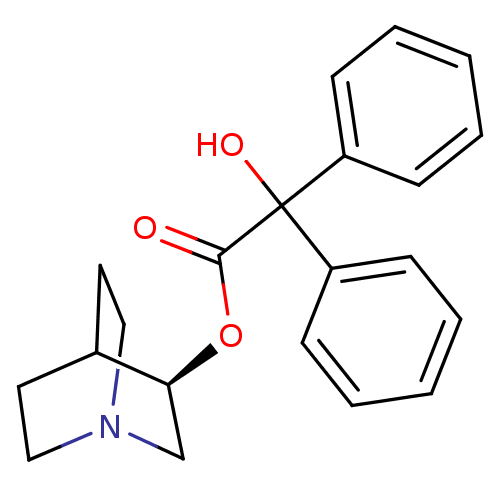 (CHEMBL558910) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | Purchase CHEMBL PC cid PC sid PDB UniChem Similars | PubMed | n/a | n/a | n/a | 0.320 | n/a | n/a | n/a | n/a | n/a |
Oak Ridge National Laboratory (ORNL) Curated by ChEMBL | Assay Description In vitro binding affinity towards Muscarinic acetylcholine receptor M1 using [3H]QNB as radioligand from rat heart tissue | J Med Chem 38: 3908-17 (1995) BindingDB Entry DOI: 10.7270/Q24M9567 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M2 (RAT) | BDBM50450592 (CHEMBL558910) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase CHEMBL PC cid PC sid PDB UniChem Similars | PubMed | n/a | n/a | n/a | 0.200 | n/a | n/a | n/a | n/a | n/a |
Oak Ridge National Laboratory (ORNL) Curated by ChEMBL | Assay Description In vitro binding affinity towards Muscarinic acetylcholine receptor M2 using [3H]-QNB as radioligand from rat heart tissue | J Med Chem 38: 3908-17 (1995) BindingDB Entry DOI: 10.7270/Q24M9567 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M2 (RAT) | BDBM50031098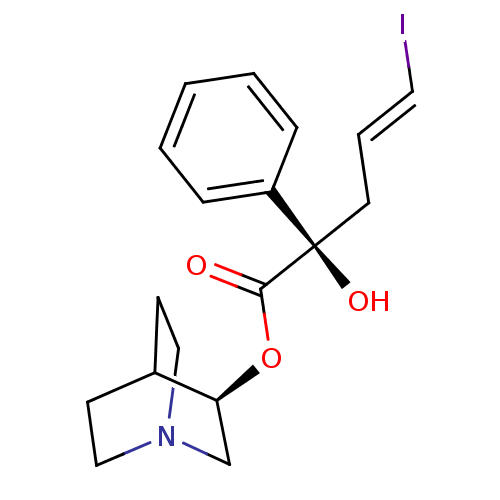 ((E)-(R)-2-Hydroxy-5-iodo-2-phenyl-pent-4-enoic aci...) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | n/a | n/a | n/a | 30 | n/a | n/a | n/a | n/a | n/a |
Oak Ridge National Laboratory (ORNL) Curated by ChEMBL | Assay Description In vitro binding affinity towards Muscarinic acetylcholine receptor M2 using [3H]-QNB as radioligand from rat heart tissue | J Med Chem 38: 3908-17 (1995) BindingDB Entry DOI: 10.7270/Q24M9567 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M4 (RAT) | BDBM50031098 ((E)-(R)-2-Hydroxy-5-iodo-2-phenyl-pent-4-enoic aci...) | PDB KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | n/a | n/a | n/a | 2.40 | n/a | n/a | n/a | n/a | n/a |
Oak Ridge National Laboratory (ORNL) Curated by ChEMBL | Assay Description In vitro binding affinity towards Muscarinic acetylcholine receptor M4 using [3H]QNB as radioligand from rat heart tissue | J Med Chem 38: 3908-17 (1995) BindingDB Entry DOI: 10.7270/Q24M9567 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M4 (RAT) | BDBM50031098 ((E)-(R)-2-Hydroxy-5-iodo-2-phenyl-pent-4-enoic aci...) | PDB KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | n/a | n/a | n/a | 0.356 | n/a | n/a | n/a | n/a | n/a |
Oak Ridge National Laboratory (ORNL) Curated by ChEMBL | Assay Description In vitro binding affinity towards Muscarinic acetylcholine receptor M4 using [3H]QNB as radioligand from rat heart tissue | J Med Chem 38: 3908-17 (1995) BindingDB Entry DOI: 10.7270/Q24M9567 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50031098 ((E)-(R)-2-Hydroxy-5-iodo-2-phenyl-pent-4-enoic aci...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | n/a | n/a | n/a | 6.80 | n/a | n/a | n/a | n/a | n/a |
Oak Ridge National Laboratory (ORNL) Curated by ChEMBL | Assay Description In vitro binding affinity towards Muscarinic acetylcholine receptor M1 using [3H]QNB as radioligand from rat heart tissue | J Med Chem 38: 3908-17 (1995) BindingDB Entry DOI: 10.7270/Q24M9567 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M2 (RAT) | BDBM50031098 ((E)-(R)-2-Hydroxy-5-iodo-2-phenyl-pent-4-enoic aci...) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | n/a | n/a | n/a | 41 | n/a | n/a | n/a | n/a | n/a |
Oak Ridge National Laboratory (ORNL) Curated by ChEMBL | Assay Description In vitro binding affinity towards Muscarinic acetylcholine receptor M2 using [3H]-QNB as radioligand from rat heart tissue | J Med Chem 38: 3908-17 (1995) BindingDB Entry DOI: 10.7270/Q24M9567 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50031098 ((E)-(R)-2-Hydroxy-5-iodo-2-phenyl-pent-4-enoic aci...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | n/a | n/a | n/a | 0.383 | n/a | n/a | n/a | n/a | n/a |
Oak Ridge National Laboratory (ORNL) Curated by ChEMBL | Assay Description In vitro binding affinity towards Muscarinic acetylcholine receptor M1 using [3H]QNB as radioligand from rat heart tissue | J Med Chem 38: 3908-17 (1995) BindingDB Entry DOI: 10.7270/Q24M9567 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (RAT) | BDBM50453895 (CHEMBL2111842) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | n/a | n/a | n/a | 0.230 | n/a | n/a | n/a | n/a | n/a |
Oak Ridge National Laboratory (ORNL) Curated by ChEMBL | Assay Description In vitro binding affinity towards Muscarinic acetylcholine receptor M1 using [3H]QNB as radioligand from rat heart tissue | J Med Chem 38: 3908-17 (1995) BindingDB Entry DOI: 10.7270/Q24M9567 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||