| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Cholinesterase |
|---|
| Ligand | BDBM8960 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1677336 (CHEMBL4027479) |
|---|
| IC50 | 7138±n/a nM |
|---|
| Citation |  Ko?ak, U; Brus, B; Knez, D; ?akelj, S; Trontelj, J; Pi?lar, A; ?ink, R; Juki?, M; ?ivin, M; Podkowa, A; Nachon, F; Brazzolotto, X; Stojan, J; Kos, J; Coquelle, N; Sa?at, K; Colletier, JP; Gobec, S The Magic of Crystal Structure-Based Inhibitor Optimization: Development of a Butyrylcholinesterase Inhibitor with Picomolar Affinity and in Vivo Activity. J Med Chem61:119-139 (2018) [PubMed] Article Ko?ak, U; Brus, B; Knez, D; ?akelj, S; Trontelj, J; Pi?lar, A; ?ink, R; Juki?, M; ?ivin, M; Podkowa, A; Nachon, F; Brazzolotto, X; Stojan, J; Kos, J; Coquelle, N; Sa?at, K; Colletier, JP; Gobec, S The Magic of Crystal Structure-Based Inhibitor Optimization: Development of a Butyrylcholinesterase Inhibitor with Picomolar Affinity and in Vivo Activity. J Med Chem61:119-139 (2018) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Cholinesterase |
|---|
| Name: | Cholinesterase |
|---|
| Synonyms: | Acylcholine acylhydrolase | Bche | Butyrylcholine esterase | Butyrylcholinesterase | CHLE_MOUSE | Choline esterase II | Pseudocholinesterase |
|---|
| Type: | Protein |
|---|
| Mol. Mass.: | 68465.99 |
|---|
| Organism: | Mus musculus (Mouse) |
|---|
| Description: | Q03311 |
|---|
| Residue: | 603 |
|---|
| Sequence: | MQTQHTKVTQTHFLLWILLLCMPFGKSHTEEDFIITTKTGRVRGLSMPVLGGTVTAFLGI
PYAQPPLGSLRFKKPQPLNKWPDIHNATQYANSCYQNIDQAFPGFQGSEMWNPNTNLSED
CLYLNVWIPVPKPKNATVMVWIYGGGFQTGTSSLPVYDGKFLARVERVIVVSMNYRVGAL
GFLAFPGNPDAPGNMGLFDQQLALQWVQRNIAAFGGNPKSITIFGESAGAASVSLHLLCP
QSYPLFTRAILESGSSNAPWAVKHPEEARNRTLTLAKFTGCSKENEMEMIKCLRSKDPQE
ILRNERFVLPSDSILSINFGPTVDGDFLTDMPHTLLQLGKVKKAQILVGVNKDEGTAFLV
YGAPGFSKDNDSLITRKEFQEGLNMYFPGVSRLGKEAVLFYYVDWLGEQSPEVYRDALDD
VIGDYNIICPALEFTKKFAELENNAFFYFFEHRSSKLPWPEWMGVMHGYEIEFVFGLPLG
RRVNYTRAEEIFSRSIMKTWANFAKYGHPNGTQGNSTMWPVFTSTEQKYLTLNTEKSKIY
SKLRAPQCQFWRLFFPKVLEMTGDIDETEQEWKAGFHRWSNYMMDWQNQFNDYTSKKESC
TAL
|
|
|
|---|
| BDBM8960 |
|---|
| n/a |
|---|
| Name | BDBM8960 |
|---|
| Synonyms: | (+/-)-2-[(1-benzylpiperidin-4-yl)methyl]-5,6-dimethoxy-indan-1-one | 2-[(1-benzylpiperidin-4-yl)methyl]-5,6-dimethoxy-2,3-dihydro-1H-inden-1-one | Aricept | Aricept odt | CHEMBL1678 | CHEMBL2337271 | CHEMBL502 | DONEPEZIL HYDROCHLORIDE | Donepezil | Donepzil | E2020 | US8999994, Donepezil | US9346818, DPH | US9586925, Donepezil | US9663465, Donepezil |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C24H29NO3 |
|---|
| Mol. Mass. | 379.492 |
|---|
| SMILES | COc1cc2CC(CC3CCN(Cc4ccccc4)CC3)C(=O)c2cc1OC |
|---|
| Structure | 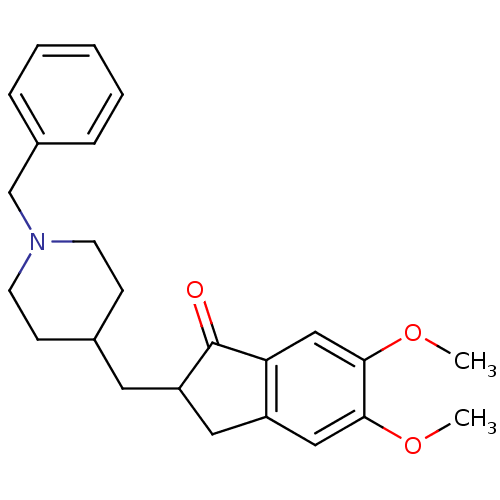
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Ko?ak, U; Brus, B; Knez, D; ?akelj, S; Trontelj, J; Pi?lar, A; ?ink, R; Juki?, M; ?ivin, M; Podkowa, A; Nachon, F; Brazzolotto, X; Stojan, J; Kos, J; Coquelle, N; Sa?at, K; Colletier, JP; Gobec, S The Magic of Crystal Structure-Based Inhibitor Optimization: Development of a Butyrylcholinesterase Inhibitor with Picomolar Affinity and in Vivo Activity. J Med Chem61:119-139 (2018) [PubMed] Article
Ko?ak, U; Brus, B; Knez, D; ?akelj, S; Trontelj, J; Pi?lar, A; ?ink, R; Juki?, M; ?ivin, M; Podkowa, A; Nachon, F; Brazzolotto, X; Stojan, J; Kos, J; Coquelle, N; Sa?at, K; Colletier, JP; Gobec, S The Magic of Crystal Structure-Based Inhibitor Optimization: Development of a Butyrylcholinesterase Inhibitor with Picomolar Affinity and in Vivo Activity. J Med Chem61:119-139 (2018) [PubMed] Article