| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Tyrosyl-DNA phosphodiesterase 2 |
|---|
| Ligand | BDBM50257100 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEBML_1689437 |
|---|
| IC50 | >111000±n/a nM |
|---|
| Citation |  Elsayed, MSA; Su, Y; Wang, P; Sethi, T; Agama, K; Ravji, A; Redon, CE; Kiselev, E; Horzmann, KA; Freeman, JL; Pommier, Y; Cushman, M Design and Synthesis of Chlorinated and Fluorinated 7-Azaindenoisoquinolines as Potent Cytotoxic Anticancer Agents That Inhibit Topoisomerase I. J Med Chem60:5364-5376 (2017) [PubMed] Article Elsayed, MSA; Su, Y; Wang, P; Sethi, T; Agama, K; Ravji, A; Redon, CE; Kiselev, E; Horzmann, KA; Freeman, JL; Pommier, Y; Cushman, M Design and Synthesis of Chlorinated and Fluorinated 7-Azaindenoisoquinolines as Potent Cytotoxic Anticancer Agents That Inhibit Topoisomerase I. J Med Chem60:5364-5376 (2017) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Tyrosyl-DNA phosphodiesterase 2 |
|---|
| Name: | Tyrosyl-DNA phosphodiesterase 2 |
|---|
| Synonyms: | EAP2 | EAPII | ETS1-associated protein 2 | ETS1-associated protein II | TDP2 | TRAF and TNF receptor-associated protein | TTRAP | TYDP2_HUMAN | Tyr-DNA phosphodiesterase 2 | Tyrosyl-DNA phosphodiesterase 2 | Tyrosyl-DNA phosphodiesterase 2 (hTDP2) | Tyrosyl-RNA phosphodiesterase | VPg unlinkase | hTDP2 |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 40916.54 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | gi_23510348 |
|---|
| Residue: | 362 |
|---|
| Sequence: | MELGSCLEGGREAAEEEGEPEVKKRRLLCVEFASVASCDAAVAQCFLAENDWEMERALNS
YFEPPVEESALERRPETISEPKTYVDLTNEETTDSTTSKISPSEDTQQENGSMFSLITWN
IDGLDLNNLSERARGVCSYLALYSPDVIFLQEVIPPYYSYLKKRSSNYEIITGHEEGYFT
AIMLKKSRVKLKSQEIIPFPSTKMMRNLLCVHVNVSGNELCLMTSHLESTRGHAAERMNQ
LKMVLKKMQEAPESATVIFAGDTNLRDREVTRCGGLPNNIVDVWEFLGKPKHCQYTWDTQ
MNSNLGITAACKLRFDRIFFRAAAEEGHIIPRSLDLLGLEKLDCGRFPSDHWGLLCNLDI
IL
|
|
|
|---|
| BDBM50257100 |
|---|
| n/a |
|---|
| Name | BDBM50257100 |
|---|
| Synonyms: | CHEMBL4079995 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C22H17FN4O3 |
|---|
| Mol. Mass. | 404.3938 |
|---|
| SMILES | COc1cnc-2c(c1)C(=O)c1c-2n(CCCn2ccnc2)c(=O)c2cc(F)ccc12 |
|---|
| Structure | 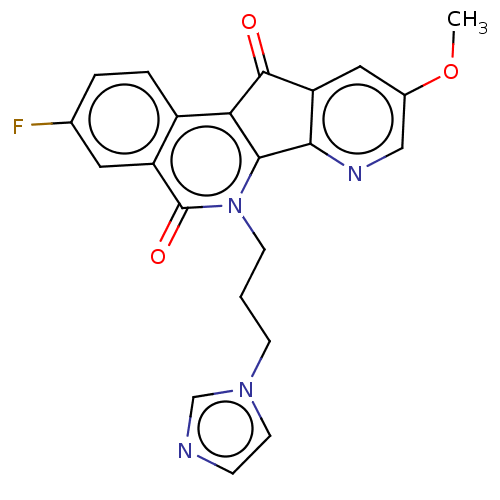
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Elsayed, MSA; Su, Y; Wang, P; Sethi, T; Agama, K; Ravji, A; Redon, CE; Kiselev, E; Horzmann, KA; Freeman, JL; Pommier, Y; Cushman, M Design and Synthesis of Chlorinated and Fluorinated 7-Azaindenoisoquinolines as Potent Cytotoxic Anticancer Agents That Inhibit Topoisomerase I. J Med Chem60:5364-5376 (2017) [PubMed] Article
Elsayed, MSA; Su, Y; Wang, P; Sethi, T; Agama, K; Ravji, A; Redon, CE; Kiselev, E; Horzmann, KA; Freeman, JL; Pommier, Y; Cushman, M Design and Synthesis of Chlorinated and Fluorinated 7-Azaindenoisoquinolines as Potent Cytotoxic Anticancer Agents That Inhibit Topoisomerase I. J Med Chem60:5364-5376 (2017) [PubMed] Article