| Citation |  Stepan, AF; Subramanyam, C; Efremov, IV; Dutra, JK; O'Sullivan, TJ; DiRico, KJ; McDonald, WS; Won, A; Dorff, PH; Nolan, CE; Becker, SL; Pustilnik, LR; Riddell, DR; Kauffman, GW; Kormos, BL; Zhang, L; Lu, Y; Capetta, SH; Green, ME; Karki, K; Sibley, E; Atchison, KP; Hallgren, AJ; Oborski, CE; Robshaw, AE; Sneed, B; O'Donnell, CJ Application of the bicyclo[1.1.1]pentane motif as a nonclassical phenyl ring bioisostere in the design of a potent and orally active ?-secretase inhibitor. J Med Chem55:3414-24 (2012) [PubMed] Article Stepan, AF; Subramanyam, C; Efremov, IV; Dutra, JK; O'Sullivan, TJ; DiRico, KJ; McDonald, WS; Won, A; Dorff, PH; Nolan, CE; Becker, SL; Pustilnik, LR; Riddell, DR; Kauffman, GW; Kormos, BL; Zhang, L; Lu, Y; Capetta, SH; Green, ME; Karki, K; Sibley, E; Atchison, KP; Hallgren, AJ; Oborski, CE; Robshaw, AE; Sneed, B; O'Donnell, CJ Application of the bicyclo[1.1.1]pentane motif as a nonclassical phenyl ring bioisostere in the design of a potent and orally active ?-secretase inhibitor. J Med Chem55:3414-24 (2012) [PubMed] Article |
|---|
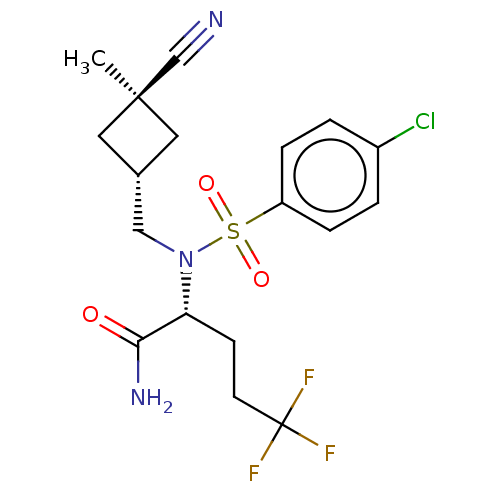
| SMILES | C[C@@]1(C[C@H](CN([C@H](CCC(F)(F)F)C(N)=O)S(=O)(=O)c2ccc(Cl)cc2)C1)C#N |r,wU:3.3,wD:6.6,1.28,(-7.4,-27.53,;-7.39,-29.08,;-7.38,-30.62,;-5.85,-30.61,;-4.5,-31.36,;-3.17,-30.59,;-1.8,-31.36,;-.47,-30.59,;.86,-31.36,;2.2,-30.58,;3.53,-31.35,;2.19,-29.04,;3.52,-29.8,;-1.8,-32.9,;-.47,-33.67,;-3.13,-33.67,;-3.18,-29.03,;-3.94,-27.7,;-4.72,-29.03,;-1.84,-28.28,;-1.81,-26.74,;-.46,-25.99,;.86,-26.78,;2.2,-26.03,;.84,-28.32,;-.51,-29.08,;-5.85,-29.08,;-8.47,-27.99,;-9.56,-26.91,)| |
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Stepan, AF; Subramanyam, C; Efremov, IV; Dutra, JK; O'Sullivan, TJ; DiRico, KJ; McDonald, WS; Won, A; Dorff, PH; Nolan, CE; Becker, SL; Pustilnik, LR; Riddell, DR; Kauffman, GW; Kormos, BL; Zhang, L; Lu, Y; Capetta, SH; Green, ME; Karki, K; Sibley, E; Atchison, KP; Hallgren, AJ; Oborski, CE; Robshaw, AE; Sneed, B; O'Donnell, CJ Application of the bicyclo[1.1.1]pentane motif as a nonclassical phenyl ring bioisostere in the design of a potent and orally active ?-secretase inhibitor. J Med Chem55:3414-24 (2012) [PubMed] Article
Stepan, AF; Subramanyam, C; Efremov, IV; Dutra, JK; O'Sullivan, TJ; DiRico, KJ; McDonald, WS; Won, A; Dorff, PH; Nolan, CE; Becker, SL; Pustilnik, LR; Riddell, DR; Kauffman, GW; Kormos, BL; Zhang, L; Lu, Y; Capetta, SH; Green, ME; Karki, K; Sibley, E; Atchison, KP; Hallgren, AJ; Oborski, CE; Robshaw, AE; Sneed, B; O'Donnell, CJ Application of the bicyclo[1.1.1]pentane motif as a nonclassical phenyl ring bioisostere in the design of a potent and orally active ?-secretase inhibitor. J Med Chem55:3414-24 (2012) [PubMed] Article