| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Serine protease 1 |
|---|
| Ligand | BDBM14320 |
|---|
| Substrate/Competitor | BDBM12679 |
|---|
| Meas. Tech. | Enzyme Assay and Determination of the Inhibition Constants |
|---|
| Ki | 600000±n/a nM |
|---|
| Citation |  McGrath, ME; Sprengeler, PA; Hirschbein, B; Somoza, JR; Lehoux, I; Janc, JW; Gjerstad, E; Graupe, M; Estiarte, A; Venkataramani, C; Liu, Y; Yee, R; Ho, JD; Green, MJ; Lee, CS; Liu, L; Tai, V; Spencer, J; Sperandio, D; Katz, BA Structure-guided design of peptide-based tryptase inhibitors. Biochemistry45:5964-73 (2006) [PubMed] Article McGrath, ME; Sprengeler, PA; Hirschbein, B; Somoza, JR; Lehoux, I; Janc, JW; Gjerstad, E; Graupe, M; Estiarte, A; Venkataramani, C; Liu, Y; Yee, R; Ho, JD; Green, MJ; Lee, CS; Liu, L; Tai, V; Spencer, J; Sperandio, D; Katz, BA Structure-guided design of peptide-based tryptase inhibitors. Biochemistry45:5964-73 (2006) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Solution Info, Assay Method |
|---|
| |
| Serine protease 1 |
|---|
| Name: | Serine protease 1 |
|---|
| Synonyms: | Beta-Trypsin | Cationic trypsin | PRSS1 | TRP1 | TRY1 | TRY1_BOVIN | TRYP1 | Trypsin | Trypsin I |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 25790.52 |
|---|
| Organism: | Bos taurus (bovine) |
|---|
| Description: | P00760 |
|---|
| Residue: | 246 |
|---|
| Sequence: | MKTFIFLALLGAAVAFPVDDDDKIVGGYTCGANTVPYQVSLNSGYHFCGGSLINSQWVVS
AAHCYKSGIQVRLGEDNINVVEGNEQFISASKSIVHPSYNSNTLNNDIMLIKLKSAASLN
SRVASISLPTSCASAGTQCLISGWGNTKSSGTSYPDVLKCLKAPILSDSSCKSAYPGQIT
SNMFCAGYLEGGKDSCQGDSGGPVVCSGKLQGIVSWGSGCAQKNKPGVYTKVCNYVSWIK
QTIASN
|
|
|
|---|
| BDBM14320 |
|---|
| BDBM12679 |
|---|
| Name | BDBM14320 |
|---|
| Synonyms: | 1-amino-isoquinoline | CHEMBL62083 | Fragment 17 | fragment 1 (J. med. chem.,50,1116) | isoquinolin-1-amine |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C9H8N2 |
|---|
| Mol. Mass. | 144.1732 |
|---|
| SMILES | Nc1nccc2ccccc12 |
|---|
| Structure | 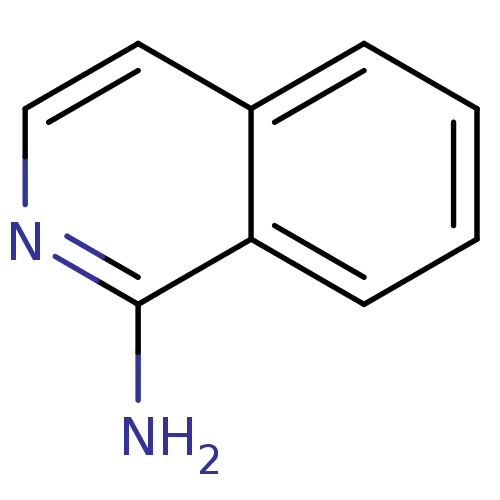
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 McGrath, ME; Sprengeler, PA; Hirschbein, B; Somoza, JR; Lehoux, I; Janc, JW; Gjerstad, E; Graupe, M; Estiarte, A; Venkataramani, C; Liu, Y; Yee, R; Ho, JD; Green, MJ; Lee, CS; Liu, L; Tai, V; Spencer, J; Sperandio, D; Katz, BA Structure-guided design of peptide-based tryptase inhibitors. Biochemistry45:5964-73 (2006) [PubMed] Article
McGrath, ME; Sprengeler, PA; Hirschbein, B; Somoza, JR; Lehoux, I; Janc, JW; Gjerstad, E; Graupe, M; Estiarte, A; Venkataramani, C; Liu, Y; Yee, R; Ho, JD; Green, MJ; Lee, CS; Liu, L; Tai, V; Spencer, J; Sperandio, D; Katz, BA Structure-guided design of peptide-based tryptase inhibitors. Biochemistry45:5964-73 (2006) [PubMed] Article