| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Peroxisome proliferator-activated receptor delta |
|---|
| Ligand | BDBM50214974 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_2051742 (CHEMBL4706441) |
|---|
| Ki | 11±n/a nM |
|---|
| Citation |  An, S; Kim, G; Kim, HJ; Ahn, S; Kim, HY; Ko, H; Hyun, YE; Nguyen, M; Jeong, J; Liu, Z; Han, J; Choi, H; Yu, J; Kim, JW; Lee, HW; Jacobson, KA; Cho, WJ; Kim, YM; Kang, KW; Noh, M; Jeong, LS Discovery and Structure-Activity Relationships of Novel Template, Truncated 1'-Homologated Adenosine Derivatives as Pure Dual PPAR?/? Modulators. J Med Chem63:16012-16027 (2020) [PubMed] Article An, S; Kim, G; Kim, HJ; Ahn, S; Kim, HY; Ko, H; Hyun, YE; Nguyen, M; Jeong, J; Liu, Z; Han, J; Choi, H; Yu, J; Kim, JW; Lee, HW; Jacobson, KA; Cho, WJ; Kim, YM; Kang, KW; Noh, M; Jeong, LS Discovery and Structure-Activity Relationships of Novel Template, Truncated 1'-Homologated Adenosine Derivatives as Pure Dual PPAR?/? Modulators. J Med Chem63:16012-16027 (2020) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Peroxisome proliferator-activated receptor delta |
|---|
| Name: | Peroxisome proliferator-activated receptor delta |
|---|
| Synonyms: | NR1C2 | NUC1 | NUCI | Nuclear hormone receptor 1 | Nuclear receptor subfamily 1 group C member 2 | PPAR delta | PPAR-beta | PPARB | PPARD | PPARD_HUMAN | Peroxisome proliferator-activated receptor | Peroxisome proliferator-activated receptor beta | Peroxisome proliferator-activated receptor delta |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 49910.45 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q03181 |
|---|
| Residue: | 441 |
|---|
| Sequence: | MEQPQEEAPEVREEEEKEEVAEAEGAPELNGGPQHALPSSSYTDLSRSSSPPSLLDQLQM
GCDGASCGSLNMECRVCGDKASGFHYGVHACEGCKGFFRRTIRMKLEYEKCERSCKIQKK
NRNKCQYCRFQKCLALGMSHNAIRFGRMPEAEKRKLVAGLTANEGSQYNPQVADLKAFSK
HIYNAYLKNFNMTKKKARSILTGKASHTAPFVIHDIETLWQAEKGLVWKQLVNGLPPYKE
ISVHVFYRCQCTTVETVRELTEFAKSIPSFSSLFLNDQVTLLKYGVHEAIFAMLASIVNK
DGLLVANGSGFVTREFLRSLRKPFSDIIEPKFEFAVKFNALELDDSDLALFIAAIILCGD
RPGLMNVPRVEAIQDTILRALEFHLQANHPDAQYLFPKLLQKMADLRQLVTEHAQMMQRI
KKTETETSLHPLLQEIYKDMY
|
|
|
|---|
| BDBM50214974 |
|---|
| n/a |
|---|
| Name | BDBM50214974 |
|---|
| Synonyms: | (2R,3R,4S)-2-(2-chloro-6-(3-iodobenzylamino)-9H-purin-9-yl)tetrahydrothiophene3,4-diol | (2R,3R,4S)-2-(6-(3-iodobenzylamino)-2-chloro-9H-purin-9-yl)-tetrahydrothiophene-3,4-diol | CHEMBL413686 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C16H15ClIN5O2S |
|---|
| Mol. Mass. | 503.745 |
|---|
| SMILES | O[C@@H]1CS[C@H]([C@@H]1O)n1cnc2c(NCc3cccc(I)c3)nc(Cl)nc12 |r| |
|---|
| Structure | 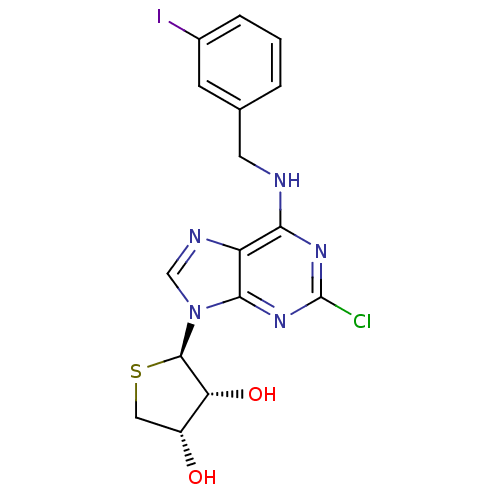
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 An, S; Kim, G; Kim, HJ; Ahn, S; Kim, HY; Ko, H; Hyun, YE; Nguyen, M; Jeong, J; Liu, Z; Han, J; Choi, H; Yu, J; Kim, JW; Lee, HW; Jacobson, KA; Cho, WJ; Kim, YM; Kang, KW; Noh, M; Jeong, LS Discovery and Structure-Activity Relationships of Novel Template, Truncated 1'-Homologated Adenosine Derivatives as Pure Dual PPAR?/? Modulators. J Med Chem63:16012-16027 (2020) [PubMed] Article
An, S; Kim, G; Kim, HJ; Ahn, S; Kim, HY; Ko, H; Hyun, YE; Nguyen, M; Jeong, J; Liu, Z; Han, J; Choi, H; Yu, J; Kim, JW; Lee, HW; Jacobson, KA; Cho, WJ; Kim, YM; Kang, KW; Noh, M; Jeong, LS Discovery and Structure-Activity Relationships of Novel Template, Truncated 1'-Homologated Adenosine Derivatives as Pure Dual PPAR?/? Modulators. J Med Chem63:16012-16027 (2020) [PubMed] Article