| Citation |  Prat, M; Fernández, D; Buil, MA; Crespo, MI; Casals, G; Ferrer, M; Tort, L; Castro, J; Monleón, JM; Gavaldà, A; Miralpeix, M; Ramos, I; Doménech, T; Vilella, D; Antón, F; Huerta, JM; Espinosa, S; López, M; Sentellas, S; González, M; Albertí, J; Segarra, V; Cárdenas, A; Beleta, J; Ryder, H Discovery of novel quaternary ammonium derivatives of (3R)-quinuclidinol esters as potent and long-acting muscarinic antagonists with potential for minimal systemic exposure after inhaled administration: identification of (3R)-3-{[hydroxy(di-2-thienyl)acetyl]oxy}-1-(3-phenoxypropyl)-1-azoniabicyclo J Med Chem52:5076-92 (2010) [PubMed] Article Prat, M; Fernández, D; Buil, MA; Crespo, MI; Casals, G; Ferrer, M; Tort, L; Castro, J; Monleón, JM; Gavaldà, A; Miralpeix, M; Ramos, I; Doménech, T; Vilella, D; Antón, F; Huerta, JM; Espinosa, S; López, M; Sentellas, S; González, M; Albertí, J; Segarra, V; Cárdenas, A; Beleta, J; Ryder, H Discovery of novel quaternary ammonium derivatives of (3R)-quinuclidinol esters as potent and long-acting muscarinic antagonists with potential for minimal systemic exposure after inhaled administration: identification of (3R)-3-{[hydroxy(di-2-thienyl)acetyl]oxy}-1-(3-phenoxypropyl)-1-azoniabicyclo J Med Chem52:5076-92 (2010) [PubMed] Article |
|---|
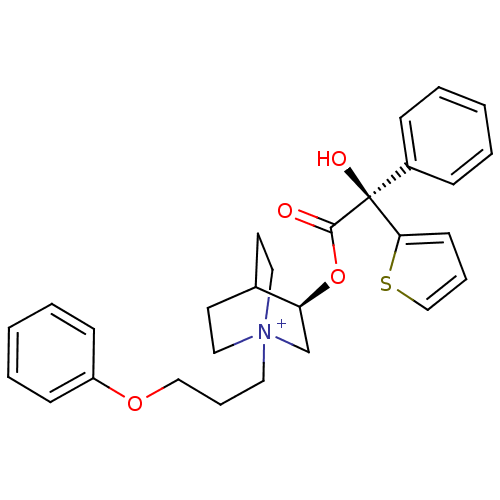
| SMILES | O[C@](C(=O)O[C@H]1C[N+]2(CCCOc3ccccc3)CCC1CC2)(c1cccs1)c1ccccc1 |r,wU:1.0,wD:5.4,1.25,(-3.92,-29.37,;-5.01,-30.46,;-3.67,-31.23,;-3.67,-32.77,;-2.34,-30.45,;-1,-31.22,;-1,-32.76,;.33,-33.52,;.31,-35.06,;1.64,-35.84,;2.98,-35.09,;4.3,-35.87,;5.65,-35.11,;6.97,-35.9,;8.31,-35.14,;8.32,-33.6,;6.99,-32.82,;5.65,-33.58,;1.65,-32.76,;1.65,-31.22,;.33,-30.44,;.75,-31.69,;-.3,-32.04,;-5.01,-28.92,;-6.26,-28.02,;-5.79,-26.55,;-4.25,-26.54,;-3.77,-28.01,;-6.34,-31.24,;-7.68,-30.47,;-9.01,-31.24,;-9,-32.78,;-7.66,-33.55,;-6.33,-32.77,)| |
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Prat, M; Fernández, D; Buil, MA; Crespo, MI; Casals, G; Ferrer, M; Tort, L; Castro, J; Monleón, JM; Gavaldà, A; Miralpeix, M; Ramos, I; Doménech, T; Vilella, D; Antón, F; Huerta, JM; Espinosa, S; López, M; Sentellas, S; González, M; Albertí, J; Segarra, V; Cárdenas, A; Beleta, J; Ryder, H Discovery of novel quaternary ammonium derivatives of (3R)-quinuclidinol esters as potent and long-acting muscarinic antagonists with potential for minimal systemic exposure after inhaled administration: identification of (3R)-3-{[hydroxy(di-2-thienyl)acetyl]oxy}-1-(3-phenoxypropyl)-1-azoniabicyclo J Med Chem52:5076-92 (2010) [PubMed] Article
Prat, M; Fernández, D; Buil, MA; Crespo, MI; Casals, G; Ferrer, M; Tort, L; Castro, J; Monleón, JM; Gavaldà, A; Miralpeix, M; Ramos, I; Doménech, T; Vilella, D; Antón, F; Huerta, JM; Espinosa, S; López, M; Sentellas, S; González, M; Albertí, J; Segarra, V; Cárdenas, A; Beleta, J; Ryder, H Discovery of novel quaternary ammonium derivatives of (3R)-quinuclidinol esters as potent and long-acting muscarinic antagonists with potential for minimal systemic exposure after inhaled administration: identification of (3R)-3-{[hydroxy(di-2-thienyl)acetyl]oxy}-1-(3-phenoxypropyl)-1-azoniabicyclo J Med Chem52:5076-92 (2010) [PubMed] Article