| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Cytochrome P450 3A4 |
|---|
| Ligand | BDBM50121391 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1518828 (CHEMBL3625502) |
|---|
| IC50 | 3000±n/a nM |
|---|
| Citation |  Hart, AC; Schroeder, GM; Wan, H; Grebinski, J; Inghrim, J; Kempson, J; Guo, J; Pitts, WJ; Tokarski, JS; Sack, JS; Khan, JA; Lippy, J; Lorenzi, MV; You, D; McDevitt, T; Vuppugalla, R; Zhang, Y; Lombardo, LJ; Trainor, GL; Purandare, AV Structure-Based Design of Selective Janus Kinase 2 Imidazo[4,5-d]pyrrolo[2,3-b]pyridine Inhibitors. ACS Med Chem Lett6:845-9 (2015) [PubMed] Article Hart, AC; Schroeder, GM; Wan, H; Grebinski, J; Inghrim, J; Kempson, J; Guo, J; Pitts, WJ; Tokarski, JS; Sack, JS; Khan, JA; Lippy, J; Lorenzi, MV; You, D; McDevitt, T; Vuppugalla, R; Zhang, Y; Lombardo, LJ; Trainor, GL; Purandare, AV Structure-Based Design of Selective Janus Kinase 2 Imidazo[4,5-d]pyrrolo[2,3-b]pyridine Inhibitors. ACS Med Chem Lett6:845-9 (2015) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Cytochrome P450 3A4 |
|---|
| Name: | Cytochrome P450 3A4 |
|---|
| Synonyms: | Albendazole monooxygenase | Albendazole sulfoxidase | CP3A4_HUMAN | CYP3A3 | CYP3A4 | CYPIIIA3 | CYPIIIA4 | Cytochrome P450 3A3 | Cytochrome P450 3A4 (CYP3A4) | Cytochrome P450 HLp | Nifedipine oxidase | Quinine 3-monooxygenase | Taurochenodeoxycholate 6-alpha-hydroxylase |
|---|
| Type: | Enzyme |
|---|
| Mol. Mass.: | 57349.57 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | n/a |
|---|
| Residue: | 503 |
|---|
| Sequence: | MALIPDLAMETWLLLAVSLVLLYLYGTHSHGLFKKLGIPGPTPLPFLGNILSYHKGFCMF
DMECHKKYGKVWGFYDGQQPVLAITDPDMIKTVLVKECYSVFTNRRPFGPVGFMKSAISI
AEDEEWKRLRSLLSPTFTSGKLKEMVPIIAQYGDVLVRNLRREAETGKPVTLKDVFGAYS
MDVITSTSFGVNIDSLNNPQDPFVENTKKLLRFDFLDPFFLSITVFPFLIPILEVLNICV
FPREVTNFLRKSVKRMKESRLEDTQKHRVDFLQLMIDSQNSKETESHKALSDLELVAQSI
IFIFAGYETTSSVLSFIMYELATHPDVQQKLQEEIDAVLPNKAPPTYDTVLQMEYLDMVV
NETLRLFPIAMRLERVCKKDVEINGMFIPKGVVVMIPSYALHRDPKYWTEPEKFLPERFS
KKNKDNIDPYIYTPFGSGPRNCIGMRFALMNMKLALIRVLQNFSFKPCKETQIPLKLSLG
GLLQPEKPVVLKVESRDGTVSGA
|
|
|
|---|
| BDBM50121391 |
|---|
| n/a |
|---|
| Name | BDBM50121391 |
|---|
| Synonyms: | CHEMBL3622137 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C23H27N7OS |
|---|
| Mol. Mass. | 449.572 |
|---|
| SMILES | CCn1c(cc2c1nc(Nc1nc(C)c(C)s1)c1ncn(C)c21)C(=O)N(C1CC1)C1CC1 |
|---|
| Structure | 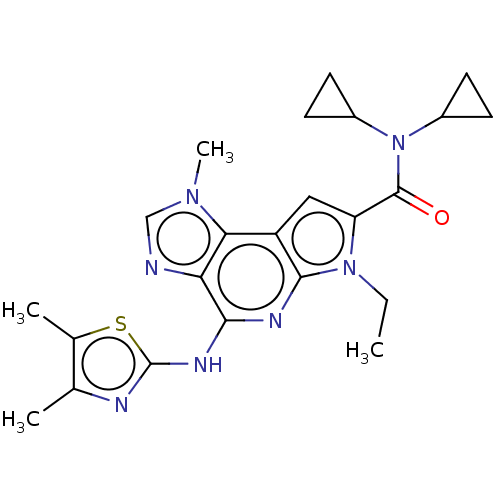
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Hart, AC; Schroeder, GM; Wan, H; Grebinski, J; Inghrim, J; Kempson, J; Guo, J; Pitts, WJ; Tokarski, JS; Sack, JS; Khan, JA; Lippy, J; Lorenzi, MV; You, D; McDevitt, T; Vuppugalla, R; Zhang, Y; Lombardo, LJ; Trainor, GL; Purandare, AV Structure-Based Design of Selective Janus Kinase 2 Imidazo[4,5-d]pyrrolo[2,3-b]pyridine Inhibitors. ACS Med Chem Lett6:845-9 (2015) [PubMed] Article
Hart, AC; Schroeder, GM; Wan, H; Grebinski, J; Inghrim, J; Kempson, J; Guo, J; Pitts, WJ; Tokarski, JS; Sack, JS; Khan, JA; Lippy, J; Lorenzi, MV; You, D; McDevitt, T; Vuppugalla, R; Zhang, Y; Lombardo, LJ; Trainor, GL; Purandare, AV Structure-Based Design of Selective Janus Kinase 2 Imidazo[4,5-d]pyrrolo[2,3-b]pyridine Inhibitors. ACS Med Chem Lett6:845-9 (2015) [PubMed] Article