

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Protease (Human immunodeficiency virus 1 (HIV-1)) | BDBM8125 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 0.0160 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Georgia State University Curated by ChEMBL | Assay Description Inhibition of HIV1 protease | J Med Chem 55: 3387-97 (2012) Article DOI: 10.1021/jm300072d BindingDB Entry DOI: 10.7270/Q2T43WZW | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Dimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A,L591M] (Human immunodeficiency virus type 1) | BDBM8125 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 0.0300 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Georgia State University | Assay Description The inhibition assays were performed in microtiter plate wells by mixing enzyme and fluorescent peptide substrate in the presence of inhibitor compou... | J Med Chem 49: 1379-87 (2006) Article DOI: 10.1021/jm050943c BindingDB Entry DOI: 10.7270/Q20C4T0V | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Alpha-1A/Alpha-1B/Alpha-1D adrenergic receptor (Rattus norvegicus (rat)-Rattus norvegicus (Rat)) | BDBM29568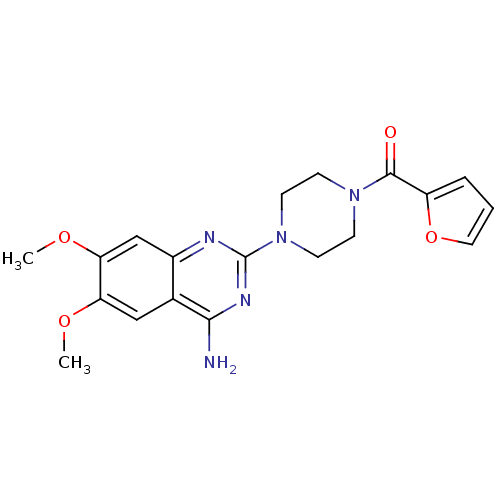 (CHEMBL2 | PRAZOSIN | PRAZOSIN HYDROCHLORIDE | [3H]...) | PDB Reactome pathway KEGG UniProtKB/SwissProt DrugBank GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE KEGG MMDB PC cid PC sid PDB UniChem Patents Similars | PubMed | 0.160 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description Compound was tested for the inhibition of [3H]prazosin binding Alpha-1 adrenergic receptor of crude rat brain membrane. | J Med Chem 25: 75-81 (1982) BindingDB Entry DOI: 10.7270/Q2TM7DBC | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A] (Human immunodeficiency virus type 1) | BDBM8125 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 0.220 | -57.3 | n/a | n/a | n/a | n/a | n/a | 5.6 | 37 |
Georgia State University | Assay Description The inhibition assays were performed in microtiter plate wells by mixing enzyme and fluorescent peptide substrate in the presence of inhibitor compou... | J Med Chem 49: 1379-87 (2006) Article DOI: 10.1021/jm050943c BindingDB Entry DOI: 10.7270/Q20C4T0V | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Protease (Human immunodeficiency virus 1 (HIV-1)) | BDBM50476647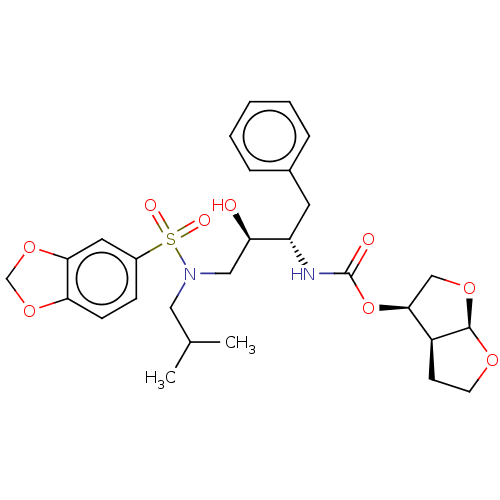 (CHEMBL178593 | GRL-98065) | PDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid PDB UniChem | PDB Article PubMed | 0.270 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Georgia State University Curated by ChEMBL | Assay Description Inhibition of wild type HIV1 protease | J Med Chem 50: 4509-15 (2007) Article DOI: 10.1021/jm070482q BindingDB Entry DOI: 10.7270/Q21G0Q1W | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Protease (Human immunodeficiency virus 1 (HIV-1)) | BDBM8125 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 0.340 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Georgia State University Curated by ChEMBL | Assay Description Inhibition of wild type HIV1 protease | J Med Chem 50: 4509-15 (2007) Article DOI: 10.1021/jm070482q BindingDB Entry DOI: 10.7270/Q21G0Q1W | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Protease (Human immunodeficiency virus 1 (HIV-1)) | BDBM31817 (GRL-02031 | methyl-2-pyrrolidinone, 19b) | PDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | MMDB PC cid PC sid PDB UniChem Patents Similars | Article PubMed | 0.440 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Georgia State University Curated by ChEMBL | Assay Description Inhibition of HIV1 wild type protease using Abz-Thr-Ile-Nle-p-nitro-Phe-Gln-Arg-NH2 as substrate after 5 mins by spectrophotometry | J Med Chem 55: 3387-97 (2012) Article DOI: 10.1021/jm300072d BindingDB Entry DOI: 10.7270/Q2T43WZW | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| HIV-1 protease (Human immunodeficiency virus) | BDBM31817 (GRL-02031 | methyl-2-pyrrolidinone, 19b) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | MMDB PC cid PC sid PDB UniChem Patents Similars | Article PubMed | 0.460 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Georgia State University Curated by ChEMBL | Assay Description Inhibition of HIV1 protease I47V mutant using Abz-Thr-Ile-Nle-p-nitro-Phe-Gln-Arg-NH2 as substrate after 5 mins by spectrophotometry | J Med Chem 55: 3387-97 (2012) Article DOI: 10.1021/jm300072d BindingDB Entry DOI: 10.7270/Q2T43WZW | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Protease (Human immunodeficiency virus 1 (HIV-1)) | BDBM50489369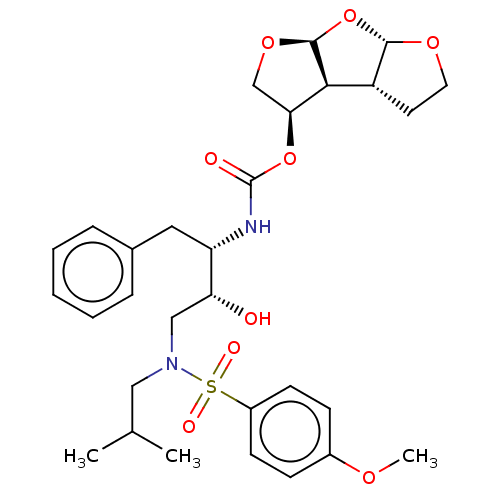 (CHEMBL1232930 | GRL-0519) | PDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid PDB UniChem | Article PubMed | 0.5 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Georgia State University Curated by ChEMBL | Assay Description Inhibition of HIV-1 wildtype protease using Abz-Thr-Ile-Nle-p-nitro-Phe-Gln-Arg-NH2 as substrate incubated for 5 mins prior to substrate addition mea... | J Med Chem 56: 1074-83 (2013) Article DOI: 10.1021/jm301519z BindingDB Entry DOI: 10.7270/Q2542RH4 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| HIV-1 protease (Human immunodeficiency virus) | BDBM8125 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 0.540 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Georgia State University Curated by ChEMBL | Assay Description Inhibition of HIV1 protease V82A mutant | J Med Chem 50: 4509-15 (2007) Article DOI: 10.1021/jm070482q BindingDB Entry DOI: 10.7270/Q21G0Q1W | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Dimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A] (Human immunodeficiency virus type 1) | BDBM517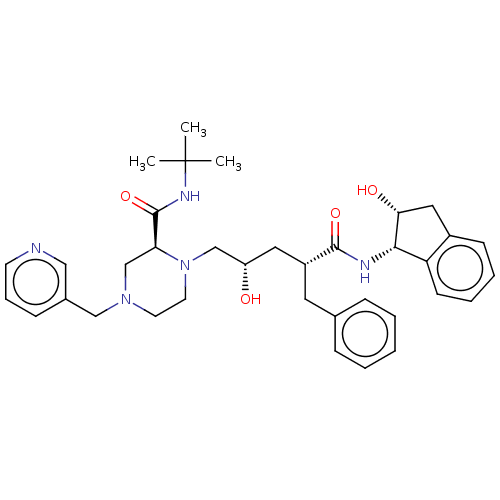 ((2S)-1-[(2S,4R)-4-benzyl-2-hydroxy-4-{[(1S,2R)-2-h...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL MCE MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 0.540 | -52.4 | n/a | n/a | n/a | n/a | n/a | 5.0 | 22 |
Georgia State University | Assay Description The Ki values were obtained from the IC50 values estimated from an inhibitor dose-response curve with the spectroscopic assay and the chromogenic sub... | J Mol Biol 354: 789-800 (2005) Article DOI: 10.1016/j.jmb.2005.09.095 BindingDB Entry DOI: 10.7270/Q2KP80CR | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Dimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A,G574S] (Human immunodeficiency virus type 1) | BDBM517 ((2S)-1-[(2S,4R)-4-benzyl-2-hydroxy-4-{[(1S,2R)-2-h...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL MCE MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 0.550 | -52.3 | n/a | n/a | n/a | n/a | n/a | 5.0 | 22 |
Georgia State University | Assay Description The Ki values were obtained from the IC50 values estimated from an inhibitor dose-response curve with the spectroscopic assay and the chromogenic sub... | J Mol Biol 354: 789-800 (2005) Article DOI: 10.1016/j.jmb.2005.09.095 BindingDB Entry DOI: 10.7270/Q2KP80CR | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| HIV-1 protease (Human immunodeficiency virus) | BDBM8125 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 0.560 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Georgia State University Curated by ChEMBL | Assay Description Inhibition of HIV1 protease I84V mutant | J Med Chem 50: 4509-15 (2007) Article DOI: 10.1021/jm070482q BindingDB Entry DOI: 10.7270/Q21G0Q1W | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Dimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A] (Human immunodeficiency virus type 1) | BDBM517 ((2S)-1-[(2S,4R)-4-benzyl-2-hydroxy-4-{[(1S,2R)-2-h...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL MCE MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 0.600 | -54.8 | n/a | n/a | n/a | n/a | n/a | 5.6 | 37 |
Georgia State University | Assay Description The inhibition assays were performed in microtiter plate wells by mixing enzyme and fluorescent peptide substrate in the presence of inhibitor compou... | J Med Chem 49: 1379-87 (2006) Article DOI: 10.1021/jm050943c BindingDB Entry DOI: 10.7270/Q20C4T0V | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| HIV-1 protease (Human immunodeficiency virus) | BDBM31817 (GRL-02031 | methyl-2-pyrrolidinone, 19b) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | MMDB PC cid PC sid PDB UniChem Patents Similars | Article PubMed | 0.650 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Georgia State University Curated by ChEMBL | Assay Description Inhibition of HIV1 protease V82A mutant using Abz-Thr-Ile-Nle-p-nitro-Phe-Gln-Arg-NH2 as substrate after 5 mins by spectrophotometry | J Med Chem 55: 3387-97 (2012) Article DOI: 10.1021/jm300072d BindingDB Entry DOI: 10.7270/Q2T43WZW | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A,L591M] (Human immunodeficiency virus type 1) | BDBM517 ((2S)-1-[(2S,4R)-4-benzyl-2-hydroxy-4-{[(1S,2R)-2-h...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL MCE MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 0.800 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Georgia State University | Assay Description The inhibition assays were performed in microtiter plate wells by mixing enzyme and fluorescent peptide substrate in the presence of inhibitor compou... | J Med Chem 49: 1379-87 (2006) Article DOI: 10.1021/jm050943c BindingDB Entry DOI: 10.7270/Q20C4T0V | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| HIV-1 protease (Human immunodeficiency virus) | BDBM31817 (GRL-02031 | methyl-2-pyrrolidinone, 19b) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | MMDB PC cid PC sid PDB UniChem Patents Similars | Article PubMed | 0.800 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Georgia State University Curated by ChEMBL | Assay Description Inhibition of HIV1 protease L76V mutant using Abz-Thr-Ile-Nle-p-nitro-Phe-Gln-Arg-NH2 as substrate after 5 mins by spectrophotometry | J Med Chem 55: 3387-97 (2012) Article DOI: 10.1021/jm300072d BindingDB Entry DOI: 10.7270/Q2T43WZW | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A,V583A] (Human immunodeficiency virus type 1) | BDBM8125 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 0.800 | -54.0 | n/a | n/a | n/a | n/a | n/a | 5.6 | 37 |
Georgia State University | Assay Description The inhibition assays were performed in microtiter plate wells by mixing enzyme and fluorescent peptide substrate in the presence of inhibitor compou... | J Med Chem 49: 1379-87 (2006) Article DOI: 10.1021/jm050943c BindingDB Entry DOI: 10.7270/Q20C4T0V | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| HIV-1 protease (Human immunodeficiency virus) | BDBM50476647 (CHEMBL178593 | GRL-98065) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | PC cid PC sid PDB UniChem | PDB Article PubMed | 0.800 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Georgia State University Curated by ChEMBL | Assay Description Inhibition of HIV1 protease V82A mutant | J Med Chem 50: 4509-15 (2007) Article DOI: 10.1021/jm070482q BindingDB Entry DOI: 10.7270/Q21G0Q1W | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| HIV-1 protease (Human immunodeficiency virus) | BDBM50476647 (CHEMBL178593 | GRL-98065) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | PC cid PC sid PDB UniChem | PDB Article PubMed | 0.850 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Georgia State University Curated by ChEMBL | Assay Description Inhibition of HIV1 protease I84V mutant | J Med Chem 50: 4509-15 (2007) Article DOI: 10.1021/jm070482q BindingDB Entry DOI: 10.7270/Q21G0Q1W | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| HIV-1 protease (Human immunodeficiency virus) | BDBM31817 (GRL-02031 | methyl-2-pyrrolidinone, 19b) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | MMDB PC cid PC sid PDB UniChem Patents Similars | Article PubMed | 0.920 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Georgia State University Curated by ChEMBL | Assay Description Inhibition of HIV1 protease N88D mutant using Abz-Thr-Ile-Nle-p-nitro-Phe-Gln-Arg-NH2 as substrate after 5 mins by spectrophotometry | J Med Chem 55: 3387-97 (2012) Article DOI: 10.1021/jm300072d BindingDB Entry DOI: 10.7270/Q2T43WZW | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A,I585V] (Human immunodeficiency virus type 1) | BDBM8125 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 1.10 | -53.2 | n/a | n/a | n/a | n/a | n/a | 5.6 | 37 |
Georgia State University | Assay Description The inhibition assays were performed in microtiter plate wells by mixing enzyme and fluorescent peptide substrate in the presence of inhibitor compou... | J Med Chem 49: 1379-87 (2006) Article DOI: 10.1021/jm050943c BindingDB Entry DOI: 10.7270/Q20C4T0V | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| HIV-1 protease (Human immunodeficiency virus) | BDBM50489369 (CHEMBL1232930 | GRL-0519) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | PC cid PC sid PDB UniChem | Article PubMed | 1.20 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Georgia State University Curated by ChEMBL | Assay Description Inhibition of HIV-1 protease R8Q mutant using Abz-Thr-Ile-Nle-p-nitro-Phe-Gln-Arg-NH2 as substrate incubated for 5 mins prior to substrate addition m... | J Med Chem 56: 1074-83 (2013) Article DOI: 10.1021/jm301519z BindingDB Entry DOI: 10.7270/Q2542RH4 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| HIV-1 protease (Human immunodeficiency virus) | BDBM8125 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 1.30 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Georgia State University Curated by ChEMBL | Assay Description Inhibition of HIV1 protease I50V mutant | J Med Chem 50: 4509-15 (2007) Article DOI: 10.1021/jm070482q BindingDB Entry DOI: 10.7270/Q21G0Q1W | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Dimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A,V583A] (Human immunodeficiency virus type 1) | BDBM517 ((2S)-1-[(2S,4R)-4-benzyl-2-hydroxy-4-{[(1S,2R)-2-h...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL MCE MMDB PC cid PC sid PDB UniChem Patents Similars | MMDB PDB Article PubMed | 1.34 | -52.7 | n/a | n/a | n/a | n/a | n/a | 5.6 | 37 |
Georgia State University | Assay Description The inhibition assays were performed in microtiter plate wells by mixing enzyme and fluorescent peptide substrate in the presence of inhibitor compou... | J Med Chem 49: 1379-87 (2006) Article DOI: 10.1021/jm050943c BindingDB Entry DOI: 10.7270/Q20C4T0V | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| D(4) dopamine receptor (RAT) | BDBM50051562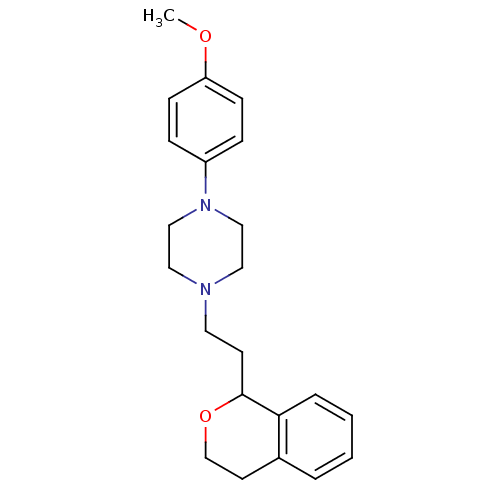 (1-(2-Isochroman-1-yl-ethyl)-4-(4-methoxy-phenyl)-p...) | Reactome pathway UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents Similars | Article PubMed | 1.40 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Pharmacia & Upjohn Curated by ChEMBL | Assay Description Binding affinity of [3H]-Spiperone towards Dopamine receptor D4 expressed in cultured cells or from rat whole brain. | J Med Chem 39: 2435-7 (1996) Article DOI: 10.1021/jm960084f BindingDB Entry DOI: 10.7270/Q2H1314Z | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A,L525I] (Human immunodeficiency virus type 1) | BDBM517 ((2S)-1-[(2S,4R)-4-benzyl-2-hydroxy-4-{[(1S,2R)-2-h...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL MCE MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 1.40 | -50.0 | n/a | n/a | n/a | n/a | n/a | 5.0 | 22 |
Georgia State University | Assay Description The Ki values were obtained from the IC50 values estimated from an inhibitor dose-response curve with the spectroscopic assay and the chromogenic sub... | J Mol Biol 354: 789-800 (2005) Article DOI: 10.1016/j.jmb.2005.09.095 BindingDB Entry DOI: 10.7270/Q2KP80CR | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Alpha-2A adrenergic receptor [16-465]/Alpha-2B adrenergic receptor/Alpha-2C adrenergic receptor (RAT-NEONATAL RAT-Rattus norvegicus (rat)) | BDBM50016897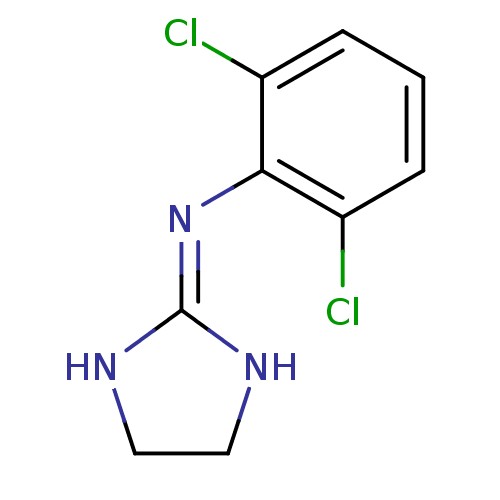 (2-(2,6-dichloroanilino)-1,3-diazacyclopentene-(2) ...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | Purchase PC cid PC sid PDB UniChem Patents Similars | PubMed | 1.70 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description Compound was tested for the inhibition of [3H]clonidine binding Alpha-2 adrenergic receptor of crude rat brain membrane | J Med Chem 25: 75-81 (1982) BindingDB Entry DOI: 10.7270/Q2TM7DBC | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| HIV-1 protease (Human immunodeficiency virus) | BDBM8125 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 1.90 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Georgia State University Curated by ChEMBL | Assay Description Inhibition of HIV1 protease D30N mutant | J Med Chem 50: 4509-15 (2007) Article DOI: 10.1021/jm070482q BindingDB Entry DOI: 10.7270/Q21G0Q1W | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Dimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A,I551V] (Human immunodeficiency virus type 1) | BDBM8125 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 2 | -51.6 | n/a | n/a | n/a | n/a | n/a | 5.6 | 37 |
Georgia State University | Assay Description The inhibition assays were performed in microtiter plate wells by mixing enzyme and fluorescent peptide substrate in the presence of inhibitor compou... | J Med Chem 49: 1379-87 (2006) Article DOI: 10.1021/jm050943c BindingDB Entry DOI: 10.7270/Q20C4T0V | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| HIV-1 protease (Human immunodeficiency virus) | BDBM50476647 (CHEMBL178593 | GRL-98065) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | PC cid PC sid PDB UniChem | PDB Article PubMed | 2.10 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Georgia State University Curated by ChEMBL | Assay Description Inhibition of HIV1 protease I50V mutant | J Med Chem 50: 4509-15 (2007) Article DOI: 10.1021/jm070482q BindingDB Entry DOI: 10.7270/Q21G0Q1W | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| D(4) dopamine receptor (RAT) | BDBM50051561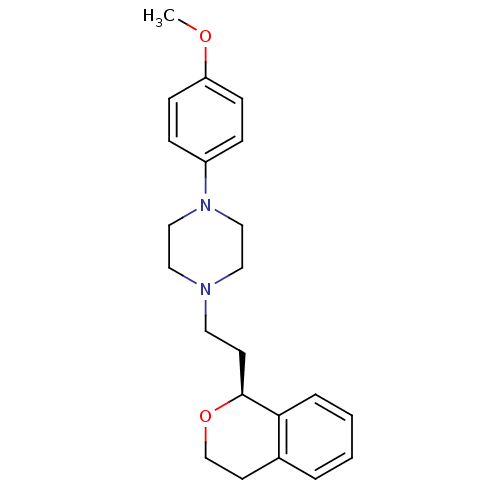 (1-((S)-2-Isochroman-1-yl-ethyl)-4-(4-methoxy-pheny...) | Reactome pathway UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 2.20 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Pharmacia & Upjohn Curated by ChEMBL | Assay Description Binding affinity of [3H]-Spiperone towards Dopamine receptor D4 expressed in cultured cells or from rat whole brain. | J Med Chem 39: 2435-7 (1996) Article DOI: 10.1021/jm960084f BindingDB Entry DOI: 10.7270/Q2H1314Z | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A,I585V] (Human immunodeficiency virus type 1) | BDBM517 ((2S)-1-[(2S,4R)-4-benzyl-2-hydroxy-4-{[(1S,2R)-2-h...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL MCE MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 2.51 | -51.1 | n/a | n/a | n/a | n/a | n/a | 5.6 | 37 |
Georgia State University | Assay Description The inhibition assays were performed in microtiter plate wells by mixing enzyme and fluorescent peptide substrate in the presence of inhibitor compou... | J Med Chem 49: 1379-87 (2006) Article DOI: 10.1021/jm050943c BindingDB Entry DOI: 10.7270/Q20C4T0V | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| D(4) dopamine receptor (RAT) | BDBM85067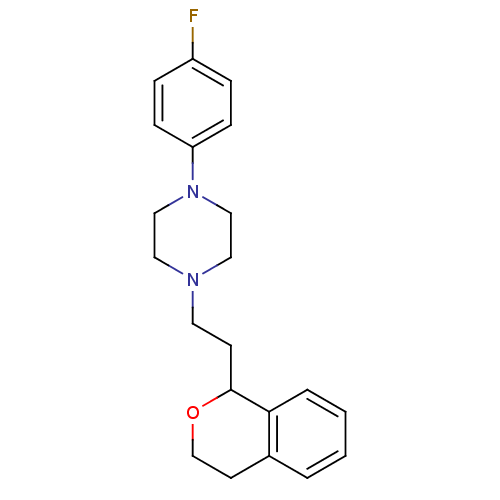 (CAS_170856-41-4 | CHEMBL81330 | PNU 96415E | PNU-9...) | Reactome pathway UniProtKB/SwissProt GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid UniChem Similars | Article PubMed | 3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Pharmacia & Upjohn Curated by ChEMBL | Assay Description Binding affinity of [3H]-Spiperone towards Dopamine receptor D4 expressed in cultured cells or from rat whole brain. | J Med Chem 39: 2435-7 (1996) Article DOI: 10.1021/jm960084f BindingDB Entry DOI: 10.7270/Q2H1314Z | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| HIV-1 protease (Human immunodeficiency virus) | BDBM50489369 (CHEMBL1232930 | GRL-0519) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | PC cid PC sid PDB UniChem | Article PubMed | 3.20 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Georgia State University Curated by ChEMBL | Assay Description Inhibition of HIV-1 protease V82A mutant using Abz-Thr-Ile-Nle-p-nitro-Phe-Gln-Arg-NH2 as substrate incubated for 5 mins prior to substrate addition ... | J Med Chem 56: 1074-83 (2013) Article DOI: 10.1021/jm301519z BindingDB Entry DOI: 10.7270/Q2542RH4 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A,L525I] (Human immunodeficiency virus type 1) | BDBM13935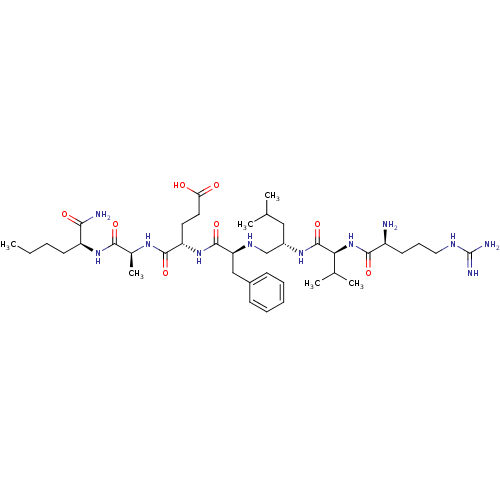 ((4S)-4-[(2S)-2-{[(2S)-2-[(2S)-2-[(2S)-2-amino-5-ca...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase MCE MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | 3.5 | -47.8 | n/a | n/a | n/a | n/a | n/a | 5.0 | 22 |
Georgia State University | Assay Description The Ki values were obtained from the IC50 values estimated from an inhibitor dose-response curve with the spectroscopic assay and the chromogenic sub... | J Mol Biol 354: 789-800 (2005) Article DOI: 10.1016/j.jmb.2005.09.095 BindingDB Entry DOI: 10.7270/Q2KP80CR | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| HIV-1 protease (Human immunodeficiency virus) | BDBM50489369 (CHEMBL1232930 | GRL-0519) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | PC cid PC sid PDB UniChem | Article PubMed | 3.5 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Georgia State University Curated by ChEMBL | Assay Description Inhibition of HIV-1 protease I54M mutant using Abz-Thr-Ile-Nle-p-nitro-Phe-Gln-Arg-NH2 as substrate incubated for 5 mins prior to substrate addition ... | J Med Chem 56: 1074-83 (2013) Article DOI: 10.1021/jm301519z BindingDB Entry DOI: 10.7270/Q2542RH4 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| D(4) dopamine receptor (RAT) | BDBM50051563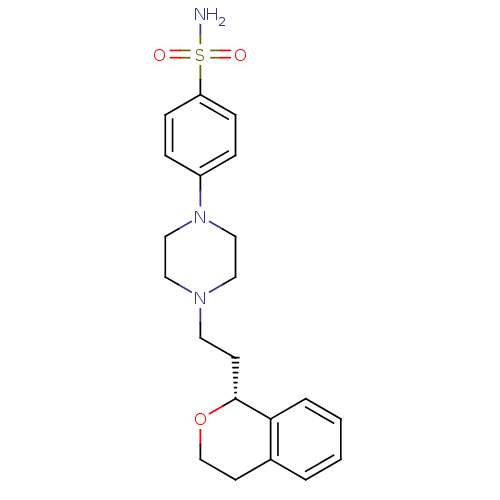 (4-[4-((R)-2-Isochroman-1-yl-ethyl)-piperazin-1-yl]...) | Reactome pathway UniProtKB/SwissProt GoogleScholar AffyNet | Purchase CHEMBL PC cid PC sid UniChem Similars | PubMed | 3.60 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Pharmacia and Upjohn, Inc. Curated by PDSP Ki Database | J Pharmacol Exp Ther 279: 1392-403 (1996) BindingDB Entry DOI: 10.7270/Q2SQ8XX9 | ||||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Alpha-1A/Alpha-1B/Alpha-1D adrenergic receptor (Rattus norvegicus (rat)-Rattus norvegicus (Rat)) | BDBM50026847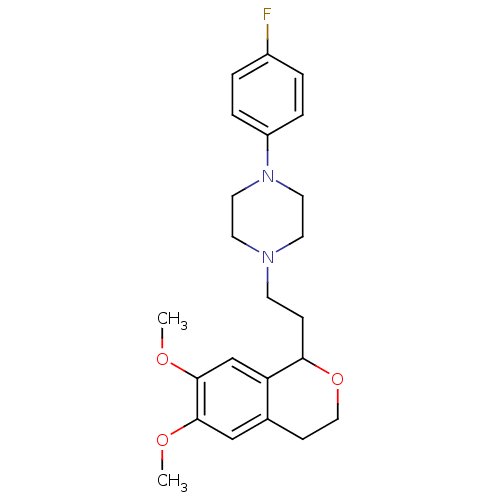 (1-[2-(6,7-Dimethoxy-isochroman-1-yl)-ethyl]-4-(4-f...) | PDB Reactome pathway KEGG UniProtKB/SwissProt DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents Similars | PubMed | 4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description Compound was tested for the inhibition of [3H]prazosin binding alpha-1-adrenergic receptor of crude rat brain membrane. | J Med Chem 25: 75-81 (1982) BindingDB Entry DOI: 10.7270/Q2TM7DBC | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Alpha-1A/Alpha-1B/Alpha-1D adrenergic receptor (Rattus norvegicus (rat)-Rattus norvegicus (Rat)) | BDBM50223655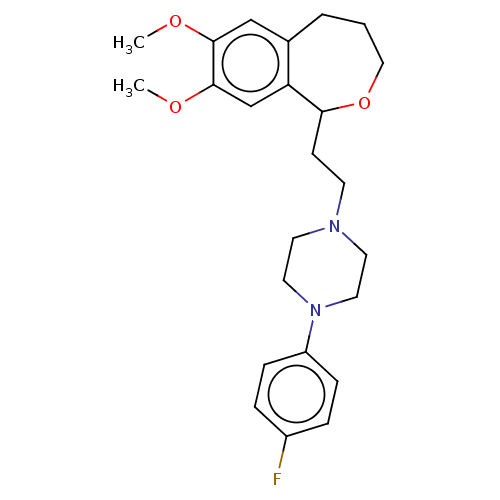 (CHEMBL160763) | PDB Reactome pathway KEGG UniProtKB/SwissProt DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | 4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description Compound was tested for the inhibition of [3H]prazosin binding Alpha-2 adrenergic receptor of crude rat brain membrane. | J Med Chem 25: 75-81 (1982) BindingDB Entry DOI: 10.7270/Q2TM7DBC | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| HIV-1 protease (Human immunodeficiency virus) | BDBM50476647 (CHEMBL178593 | GRL-98065) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | PC cid PC sid PDB UniChem | PDB Article PubMed | 4.60 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Georgia State University Curated by ChEMBL | Assay Description Inhibition of HIV1 protease D30N mutant | J Med Chem 50: 4509-15 (2007) Article DOI: 10.1021/jm070482q BindingDB Entry DOI: 10.7270/Q21G0Q1W | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Dimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A,D531N] (Human immunodeficiency virus type 1) | BDBM8125 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 6.60 | -48.6 | n/a | n/a | n/a | n/a | n/a | 5.6 | 37 |
Georgia State University | Assay Description The inhibition assays were performed in microtiter plate wells by mixing enzyme and fluorescent peptide substrate in the presence of inhibitor compou... | J Med Chem 49: 1379-87 (2006) Article DOI: 10.1021/jm050943c BindingDB Entry DOI: 10.7270/Q20C4T0V | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Dimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A,D531N] (Human immunodeficiency virus type 1) | BDBM517 ((2S)-1-[(2S,4R)-4-benzyl-2-hydroxy-4-{[(1S,2R)-2-h...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL MCE MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 7 | -48.4 | n/a | n/a | n/a | n/a | n/a | 5.6 | 37 |
Georgia State University | Assay Description The inhibition assays were performed in microtiter plate wells by mixing enzyme and fluorescent peptide substrate in the presence of inhibitor compou... | J Med Chem 49: 1379-87 (2006) Article DOI: 10.1021/jm050943c BindingDB Entry DOI: 10.7270/Q20C4T0V | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| D(4) dopamine receptor (RAT) | BDBM50051565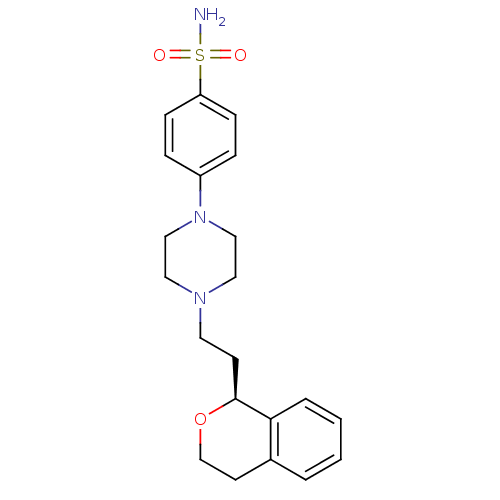 (4-[4-((S)-2-Isochroman-1-yl-ethyl)-piperazin-1-yl]...) | Reactome pathway UniProtKB/SwissProt GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid UniChem Similars | Article PubMed | 7.20 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Pharmacia & Upjohn Curated by ChEMBL | Assay Description Binding affinity of [3H]-Spiperone towards Dopamine receptor D4 expressed in cultured cells or from rat whole brain. | J Med Chem 39: 2435-7 (1996) Article DOI: 10.1021/jm960084f BindingDB Entry DOI: 10.7270/Q2H1314Z | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| HIV-1 protease (Human immunodeficiency virus) | BDBM50489369 (CHEMBL1232930 | GRL-0519) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | PC cid PC sid PDB UniChem | Article PubMed | 8.90 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Georgia State University Curated by ChEMBL | Assay Description Inhibition of HIV-1 protease D30N mutant using Abz-Thr-Ile-Nle-p-nitro-Phe-Gln-Arg-NH2 as substrate incubated for 5 mins prior to substrate addition ... | J Med Chem 56: 1074-83 (2013) Article DOI: 10.1021/jm301519z BindingDB Entry DOI: 10.7270/Q2542RH4 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| D(4) dopamine receptor (RAT) | BDBM50001884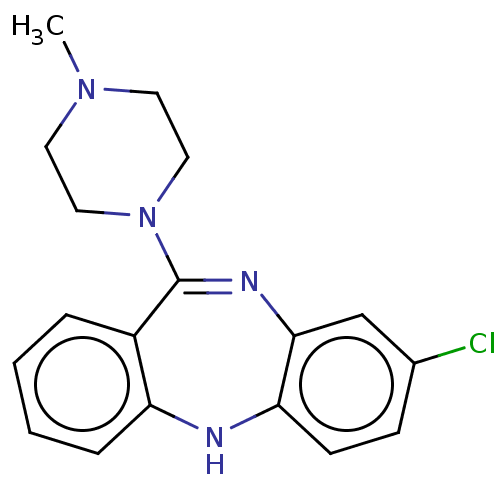 (2-[4-(4-Methyl-benzyl)-piperazin-1-yl]-1-(2-methyl...) | Reactome pathway UniProtKB/SwissProt GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE KEGG PC cid PC sid PDB UniChem Patents Similars | Article PubMed | 9 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Pharmacia & Upjohn Curated by ChEMBL | Assay Description Binding affinity of [3H]-spiperone towards cloned mammalian Dopamine receptor D4 expressed in cultured cells or from rat whole brain | J Med Chem 39: 2435-7 (1996) Article DOI: 10.1021/jm960084f BindingDB Entry DOI: 10.7270/Q2H1314Z | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Alpha-2A adrenergic receptor [16-465]/Alpha-2B adrenergic receptor/Alpha-2C adrenergic receptor (RAT-NEONATAL RAT-Rattus norvegicus (rat)) | BDBM31046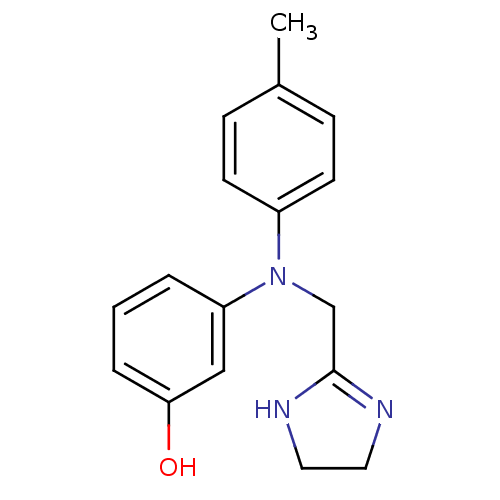 (3-[4,5-dihydro-1H-imidazol-2-ylmethyl-(4-methylphe...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | Purchase DrugBank MCE PC cid PC sid UniChem Similars | PubMed | 9.30 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description Compound was tested for the inhibition of [3H]clonidine binding Alpha-2 adrenergic receptor of crude rat brain membrane | J Med Chem 25: 75-81 (1982) BindingDB Entry DOI: 10.7270/Q2TM7DBC | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Alpha-1A/Alpha-1B/Alpha-1D adrenergic receptor (Rattus norvegicus (rat)-Rattus norvegicus (Rat)) | BDBM31046 (3-[4,5-dihydro-1H-imidazol-2-ylmethyl-(4-methylphe...) | PDB Reactome pathway KEGG UniProtKB/SwissProt DrugBank GoogleScholar AffyNet | Purchase DrugBank MCE PC cid PC sid UniChem Similars | PubMed | 9.80 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description Compound was tested for the inhibition of [3H]prazosin binding Alpha-1 adrenergic receptor of crude rat brain membrane. | J Med Chem 25: 75-81 (1982) BindingDB Entry DOI: 10.7270/Q2TM7DBC | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| D(4) dopamine receptor (Homo sapiens (Human)) | BDBM50051563 (4-[4-((R)-2-Isochroman-1-yl-ethyl)-piperazin-1-yl]...) | PDB Reactome pathway KEGG UniProtKB/SwissProt DrugBank antibodypedia GoogleScholar AffyNet | Purchase CHEMBL PC cid PC sid UniChem Similars | PubMed | 10.1 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Pharmacia and Upjohn, Inc. Curated by PDSP Ki Database | J Pharmacol Exp Ther 279: 1392-403 (1996) BindingDB Entry DOI: 10.7270/Q2SQ8XX9 | ||||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [501-599,Q508K,L534I,L564I,C568A,C596A,I551V] (Human immunodeficiency virus type 1) | BDBM517 ((2S)-1-[(2S,4R)-4-benzyl-2-hydroxy-4-{[(1S,2R)-2-h...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL MCE MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 10.4 | -47.4 | n/a | n/a | n/a | n/a | n/a | 5.6 | 37 |
Georgia State University | Assay Description The inhibition assays were performed in microtiter plate wells by mixing enzyme and fluorescent peptide substrate in the presence of inhibitor compou... | J Med Chem 49: 1379-87 (2006) Article DOI: 10.1021/jm050943c BindingDB Entry DOI: 10.7270/Q20C4T0V | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Displayed 1 to 50 (of 334 total ) | Next | Last >> |