

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Peptide deformylase (Escherichia coli) | BDBM50153088 (CHEMBL365416 | N-((3S,6R)-3-tert-Butyl-2,5-dioxo-1...) | PDB MMDB KEGG UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 0.220 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibitory effect against E. coli peptide deformylase (PDF) by AAP assay | J Med Chem 47: 4941-9 (2004) Article DOI: 10.1021/jm049592c BindingDB Entry DOI: 10.7270/Q2WD401S | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Peptide deformylase (Escherichia coli) | BDBM50153085 (CHEMBL364836 | N-((3S,6R)-3-tert-Butyl-2,5-dioxo-1...) | PDB MMDB KEGG UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 0.230 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibitory effect against E. coli peptide deformylase (PDF) by AAP assay | J Med Chem 47: 4941-9 (2004) Article DOI: 10.1021/jm049592c BindingDB Entry DOI: 10.7270/Q2WD401S | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Peptide deformylase (Escherichia coli) | BDBM50153083 (CHEMBL441502 | N-((3S,6R)-3-tert-Butyl-2,5-dioxo-1...) | PDB MMDB KEGG UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 0.330 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibitory effect against E. coli peptide deformylase (PDF) by AAP assay | J Med Chem 47: 4941-9 (2004) Article DOI: 10.1021/jm049592c BindingDB Entry DOI: 10.7270/Q2WD401S | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Peptide deformylase (Escherichia coli) | BDBM50131892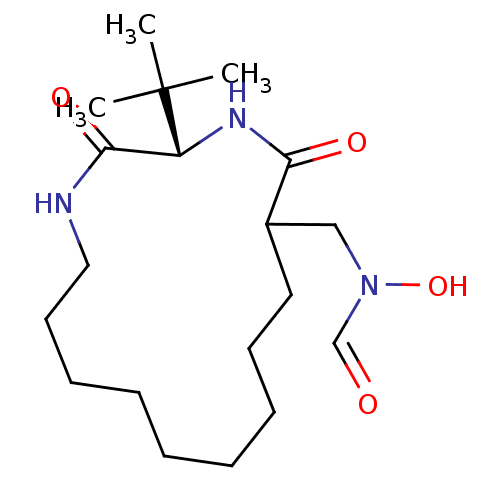 (CHEMBL421252 | N-(3-tert-Butyl-2,5-dioxo-1,4diaza-...) | PDB MMDB KEGG UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 0.670 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Binding affinity against Co(II)-substituted E. coli peptide deformylase was determined | J Med Chem 46: 3771-4 (2003) Article DOI: 10.1021/jm034113f BindingDB Entry DOI: 10.7270/Q2765DQ4 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Peptide deformylase (Escherichia coli) | BDBM50153086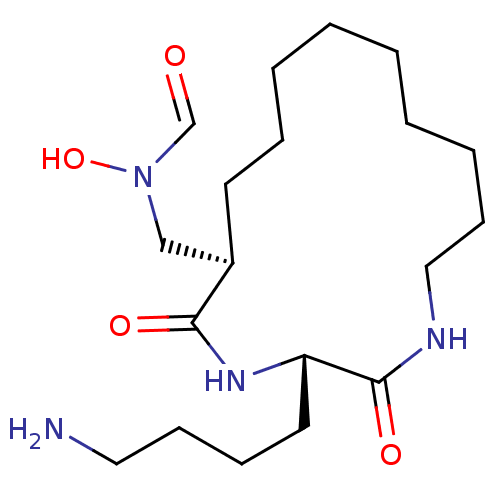 (CHEMBL188671 | N-[(1R,2S)-3-(4-Amino-butyl)-2,5-di...) | PDB MMDB KEGG UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibitory effect against E. coli peptide deformylase (PDF) by AAP assay | J Med Chem 47: 4941-9 (2004) Article DOI: 10.1021/jm049592c BindingDB Entry DOI: 10.7270/Q2WD401S | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Peptide deformylase (Escherichia coli) | BDBM50104501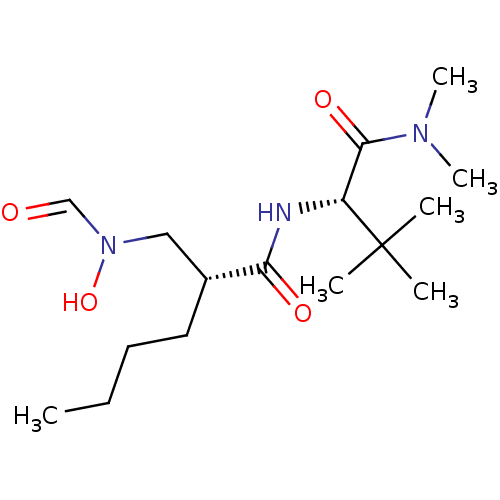 ((R)-2-[(Formyl-hydroxy-amino)-methyl]-hexanoic aci...) | PDB MMDB KEGG UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | CHEMBL DrugBank MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 11 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibitory effect against E. coli peptide deformylase (PDF) by DPPI assay | J Med Chem 47: 4941-9 (2004) Article DOI: 10.1021/jm049592c BindingDB Entry DOI: 10.7270/Q2WD401S | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Peptide deformylase (Escherichia coli) | BDBM50153084 (CHEMBL361449 | N-(4-{(3R,14S)-14-[(Formyl-hydroxy-...) | PDB MMDB KEGG UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 12 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibitory effect against E. coli peptide deformylase (PDF) by AAP assay | J Med Chem 47: 4941-9 (2004) Article DOI: 10.1021/jm049592c BindingDB Entry DOI: 10.7270/Q2WD401S | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Peptide deformylase (Escherichia coli) | BDBM50153087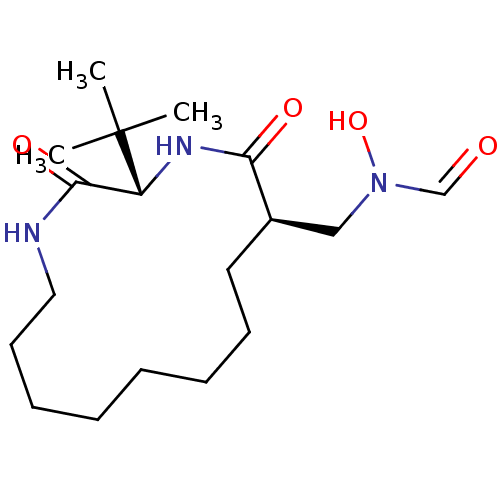 (CHEMBL188894 | N-((3S,6R)-3-tert-Butyl-2,5-dioxo-1...) | PDB MMDB KEGG UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 13.7 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibitory effect against E. coli peptide deformylase (PDF) by AAP assay | J Med Chem 47: 4941-9 (2004) Article DOI: 10.1021/jm049592c BindingDB Entry DOI: 10.7270/Q2WD401S | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Peptide deformylase (Escherichia coli) | BDBM50153080 ((R)-2-[(Formyl-hydroxy-amino)-methyl]-hexanoic aci...) | PDB MMDB KEGG UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents | Article PubMed | 23 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibitory effect against E. coli peptide deformylase (PDF) by DPPI assay | J Med Chem 47: 4941-9 (2004) Article DOI: 10.1021/jm049592c BindingDB Entry DOI: 10.7270/Q2WD401S | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Peptide deformylase (Escherichia coli) | BDBM50153081 (CHEMBL365910 | N-((3S,6R)-3-tert-Butyl-2,5-dioxo-1...) | PDB MMDB KEGG UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 25 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibitory effect against E. coli peptide deformylase (PDF) by AAP assay | J Med Chem 47: 4941-9 (2004) Article DOI: 10.1021/jm049592c BindingDB Entry DOI: 10.7270/Q2WD401S | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Peptide deformylase (Escherichia coli) | BDBM50153079 (CHEMBL364979 | N-[(3S,6R)-3-(4-Amino-butyl)-2,5-di...) | PDB MMDB KEGG UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 36 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibitory effect against E. coli peptide deformylase (PDF) by AAP assay | J Med Chem 47: 4941-9 (2004) Article DOI: 10.1021/jm049592c BindingDB Entry DOI: 10.7270/Q2WD401S | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Peptide deformylase (Escherichia coli) | BDBM50153087 (CHEMBL188894 | N-((3S,6R)-3-tert-Butyl-2,5-dioxo-1...) | PDB MMDB KEGG UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 63 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibitory effect against E. coli peptide deformylase (PDF) by DPPI assay | J Med Chem 47: 4941-9 (2004) Article DOI: 10.1021/jm049592c BindingDB Entry DOI: 10.7270/Q2WD401S | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Peptide deformylase (Escherichia coli) | BDBM50153082 ((R)-2-[(Formyl-hydroxy-amino)-methyl]-heptanoic ac...) | PDB MMDB KEGG UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents Similars | Article PubMed | 74 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibitory effect against E. coli peptide deformylase (PDF) by DPPI assay | J Med Chem 47: 4941-9 (2004) Article DOI: 10.1021/jm049592c BindingDB Entry DOI: 10.7270/Q2WD401S | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Peptide deformylase (Escherichia coli) | BDBM50153088 (CHEMBL365416 | N-((3S,6R)-3-tert-Butyl-2,5-dioxo-1...) | PDB MMDB KEGG UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 77 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibitory effect against E. coli peptide deformylase (PDF) by DPPI assay | J Med Chem 47: 4941-9 (2004) Article DOI: 10.1021/jm049592c BindingDB Entry DOI: 10.7270/Q2WD401S | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Peptide deformylase (Escherichia coli) | BDBM50153086 (CHEMBL188671 | N-[(1R,2S)-3-(4-Amino-butyl)-2,5-di...) | PDB MMDB KEGG UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 92 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibitory effect against E. coli peptide deformylase (PDF) by DPPI assay | J Med Chem 47: 4941-9 (2004) Article DOI: 10.1021/jm049592c BindingDB Entry DOI: 10.7270/Q2WD401S | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Peptide deformylase (Escherichia coli) | BDBM50153085 (CHEMBL364836 | N-((3S,6R)-3-tert-Butyl-2,5-dioxo-1...) | PDB MMDB KEGG UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 96 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibitory effect against E. coli peptide deformylase (PDF) by DPPI assay | J Med Chem 47: 4941-9 (2004) Article DOI: 10.1021/jm049592c BindingDB Entry DOI: 10.7270/Q2WD401S | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Peptide deformylase (Escherichia coli) | BDBM50153083 (CHEMBL441502 | N-((3S,6R)-3-tert-Butyl-2,5-dioxo-1...) | PDB MMDB KEGG UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 109 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibitory effect against E. coli peptide deformylase (PDF) by DPPI assay | J Med Chem 47: 4941-9 (2004) Article DOI: 10.1021/jm049592c BindingDB Entry DOI: 10.7270/Q2WD401S | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Peptide deformylase (Escherichia coli) | BDBM50153079 (CHEMBL364979 | N-[(3S,6R)-3-(4-Amino-butyl)-2,5-di...) | PDB MMDB KEGG UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 258 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibitory effect against E. coli peptide deformylase (PDF) by DPPI assay | J Med Chem 47: 4941-9 (2004) Article DOI: 10.1021/jm049592c BindingDB Entry DOI: 10.7270/Q2WD401S | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| S-ribosylhomocysteine lyase (Bacillus subtilis) | BDBM50186746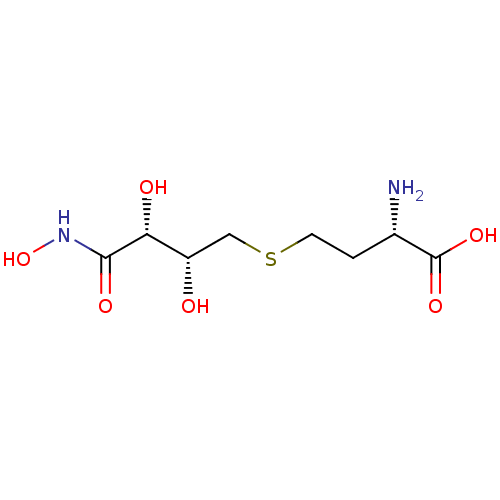 ((2S)-2-AMINO-4-[(2R,3R)-2,3-DIHYDROXY-3-N-HYDROXYC...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | 370 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibition of cobalt substituted Bacillus subtilis LuxS | J Med Chem 49: 3003-11 (2006) Article DOI: 10.1021/jm060047g BindingDB Entry DOI: 10.7270/Q25X28JQ | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| S-ribosylhomocysteine lyase (Bacillus subtilis) | BDBM50186746 ((2S)-2-AMINO-4-[(2R,3R)-2,3-DIHYDROXY-3-N-HYDROXYC...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | 430 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibition of ferrous substituted Bacillus subtilis LuxS | J Med Chem 49: 3003-11 (2006) Article DOI: 10.1021/jm060047g BindingDB Entry DOI: 10.7270/Q25X28JQ | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| S-ribosylhomocysteine lyase (Bacillus subtilis) | BDBM50304165 (CHEMBL593446 | S-(5-Deoxy-D-xylofuranos-5-yl)-L-ho...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 430 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Florida International University Curated by ChEMBL | Assay Description Inhibition of Bacillus subtilis LuxS assessed as enzyme-inhibitor dissociation constant | Bioorg Med Chem 17: 6699-706 (2009) Article DOI: 10.1016/j.bmc.2009.07.057 BindingDB Entry DOI: 10.7270/Q2PZ58XS | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (docked) | ||||||||||||
| S-ribosylhomocysteine lyase (Bacillus subtilis) | BDBM50304168 ((2S)-2-amino-4-(((2S,3S,4S)-3-fluoro-4,5-dihydroxy...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 700 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Florida International University Curated by ChEMBL | Assay Description Inhibition of Bacillus subtilis LuxS assessed as enzyme-inhibitor dissociation constant | Bioorg Med Chem 17: 6699-706 (2009) Article DOI: 10.1016/j.bmc.2009.07.057 BindingDB Entry DOI: 10.7270/Q2PZ58XS | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Peptide deformylase (Escherichia coli) | BDBM50153084 (CHEMBL361449 | N-(4-{(3R,14S)-14-[(Formyl-hydroxy-...) | PDB MMDB KEGG UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 710 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibitory effect against E. coli peptide deformylase (PDF) by DPPI assay | J Med Chem 47: 4941-9 (2004) Article DOI: 10.1021/jm049592c BindingDB Entry DOI: 10.7270/Q2WD401S | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| S-ribosylhomocysteine lyase (Bacillus subtilis) | BDBM50186747 ((2S)-2-AMINO-4-[(2R,3S)-2,3-DIHYDROXY-3-N-HYDROXYC...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | 720 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibition of ferrous substituted Bacillus subtilis LuxS | J Med Chem 49: 3003-11 (2006) Article DOI: 10.1021/jm060047g BindingDB Entry DOI: 10.7270/Q25X28JQ | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| S-ribosylhomocysteine lyase (Bacillus subtilis) | BDBM50186747 ((2S)-2-AMINO-4-[(2R,3S)-2,3-DIHYDROXY-3-N-HYDROXYC...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | 720 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibition of cobalt substituted Bacillus subtilis LuxS | J Med Chem 49: 3003-11 (2006) Article DOI: 10.1021/jm060047g BindingDB Entry DOI: 10.7270/Q25X28JQ | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| S-ribosylhomocysteine lyase (Bacillus subtilis) | BDBM50304169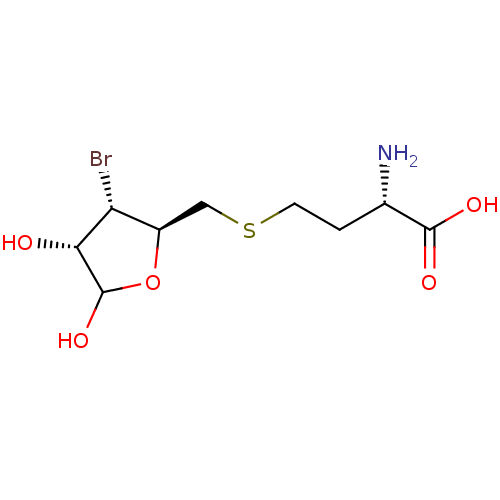 ((2S)-2-amino-4-(((2R,3S,4S)-3-bromo-4,5-dihydroxyt...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem | Article PubMed | 1.40E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Florida International University Curated by ChEMBL | Assay Description Inhibition of Bacillus subtilis LuxS assessed as enzyme-inhibitor dissociation constant | Bioorg Med Chem 17: 6699-706 (2009) Article DOI: 10.1016/j.bmc.2009.07.057 BindingDB Entry DOI: 10.7270/Q2PZ58XS | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Peptide deformylase (Escherichia coli) | BDBM50153081 (CHEMBL365910 | N-((3S,6R)-3-tert-Butyl-2,5-dioxo-1...) | PDB MMDB KEGG UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 2.10E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibitory effect against E. coli peptide deformylase (PDF) by DPPI assay | J Med Chem 47: 4941-9 (2004) Article DOI: 10.1021/jm049592c BindingDB Entry DOI: 10.7270/Q2WD401S | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| S-ribosylhomocysteine lyase (Bacillus subtilis) | BDBM50304166 (CHEMBL593447 | S-(3,5-Dideoxy-3-fluoro-D-xylofuran...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 2.80E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Florida International University Curated by ChEMBL | Assay Description Inhibition of Bacillus subtilis LuxS assessed as enzyme-inhibitor dissociation constant | Bioorg Med Chem 17: 6699-706 (2009) Article DOI: 10.1016/j.bmc.2009.07.057 BindingDB Entry DOI: 10.7270/Q2PZ58XS | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Tyrosine-protein phosphatase non-receptor type 1 (Homo sapiens (Human)) | BDBM50119679 (2-[(S)-2-(2-{2-[4-(2-Bromo-acetyl)-phenoxy]-acetyl...) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | 2.80E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Dissociation constant of the compound towards Protein-tyrosine phosphatase 1B receptor-inhibitor complex was determined using PNP as substrate | Bioorg Med Chem Lett 12: 3047-50 (2002) BindingDB Entry DOI: 10.7270/Q2FT8KCX | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| S-ribosylhomocysteine lyase (Escherichia coli (strain K12)) | BDBM50186747 ((2S)-2-AMINO-4-[(2R,3S)-2,3-DIHYDROXY-3-N-HYDROXYC...) | PDB KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | 3.20E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibition of cobalt substituted Escherichia coli LuxS | J Med Chem 49: 3003-11 (2006) Article DOI: 10.1021/jm060047g BindingDB Entry DOI: 10.7270/Q25X28JQ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| S-ribosylhomocysteine lyase (Bacillus subtilis) | BDBM50304165 (CHEMBL593446 | S-(5-Deoxy-D-xylofuranos-5-yl)-L-ho...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 4.20E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Florida International University Curated by ChEMBL | Assay Description Inhibition of Bacillus subtilis LuxS | Bioorg Med Chem 17: 6699-706 (2009) Article DOI: 10.1016/j.bmc.2009.07.057 BindingDB Entry DOI: 10.7270/Q2PZ58XS | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (docked) | ||||||||||||
| S-ribosylhomocysteine lyase (Bacillus subtilis) | BDBM50304166 (CHEMBL593447 | S-(3,5-Dideoxy-3-fluoro-D-xylofuran...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 7.70E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Florida International University Curated by ChEMBL | Assay Description Inhibition of Bacillus subtilis LuxS | Bioorg Med Chem 17: 6699-706 (2009) Article DOI: 10.1016/j.bmc.2009.07.057 BindingDB Entry DOI: 10.7270/Q2PZ58XS | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| S-ribosylhomocysteine lyase (Bacillus subtilis) | BDBM50304169 ((2S)-2-amino-4-(((2R,3S,4S)-3-bromo-4,5-dihydroxyt...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem | Article PubMed | 7.90E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Florida International University Curated by ChEMBL | Assay Description Inhibition of Bacillus subtilis LuxS | Bioorg Med Chem 17: 6699-706 (2009) Article DOI: 10.1016/j.bmc.2009.07.057 BindingDB Entry DOI: 10.7270/Q2PZ58XS | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| S-ribosylhomocysteine lyase (Bacillus subtilis) | BDBM50304167 (CHEMBL610483 | S-(3,5-Dideoxy-3-fluoro-1-O-methyl-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 8.80E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Florida International University Curated by ChEMBL | Assay Description Inhibition of Bacillus subtilis LuxS assessed as enzyme-inhibitor dissociation constant | Bioorg Med Chem 17: 6699-706 (2009) Article DOI: 10.1016/j.bmc.2009.07.057 BindingDB Entry DOI: 10.7270/Q2PZ58XS | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| S-ribosylhomocysteine lyase (Vibrio harveyi (strain ATCC BAA-1116 / BB120)) | BDBM50186747 ((2S)-2-AMINO-4-[(2R,3S)-2,3-DIHYDROXY-3-N-HYDROXYC...) | KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | 9.70E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibition of cobalt substituted Vibrio harveyi LuxS | J Med Chem 49: 3003-11 (2006) Article DOI: 10.1021/jm060047g BindingDB Entry DOI: 10.7270/Q25X28JQ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Tyrosine-protein phosphatase non-receptor type 1 (Homo sapiens (Human)) | BDBM50119683 (2-[(S)-2-(2-{[4'-(2-Bromo-acetyl)-biphenyl-2-carbo...) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem | PubMed | 9.90E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Dissociation constant of the compound towards Protein-tyrosine phosphatase 1B receptor-inhibitor complex was determined using PNP as substrate | Bioorg Med Chem Lett 12: 3047-50 (2002) BindingDB Entry DOI: 10.7270/Q2FT8KCX | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| S-ribosylhomocysteine lyase (Bacillus subtilis) | BDBM50304168 ((2S)-2-amino-4-(((2S,3S,4S)-3-fluoro-4,5-dihydroxy...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 1.06E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Florida International University Curated by ChEMBL | Assay Description Inhibition of Bacillus subtilis LuxS | Bioorg Med Chem 17: 6699-706 (2009) Article DOI: 10.1016/j.bmc.2009.07.057 BindingDB Entry DOI: 10.7270/Q2PZ58XS | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| S-ribosylhomocysteine lyase (Bacillus subtilis) | BDBM50186746 ((2S)-2-AMINO-4-[(2R,3R)-2,3-DIHYDROXY-3-N-HYDROXYC...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | 1.06E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibition of zinc substituted Bacillus subtilis LuxS | J Med Chem 49: 3003-11 (2006) Article DOI: 10.1021/jm060047g BindingDB Entry DOI: 10.7270/Q25X28JQ | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| S-ribosylhomocysteine lyase (Escherichia coli (strain K12)) | BDBM50186746 ((2S)-2-AMINO-4-[(2R,3R)-2,3-DIHYDROXY-3-N-HYDROXYC...) | PDB KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | 1.27E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibition of cobalt substituted Escherichia coli LuxS | J Med Chem 49: 3003-11 (2006) Article DOI: 10.1021/jm060047g BindingDB Entry DOI: 10.7270/Q25X28JQ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| S-ribosylhomocysteine lyase (Vibrio harveyi (strain ATCC BAA-1116 / BB120)) | BDBM50186746 ((2S)-2-AMINO-4-[(2R,3R)-2,3-DIHYDROXY-3-N-HYDROXYC...) | KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | 1.28E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibition of cobalt substituted Vibrio harveyi LuxS | J Med Chem 49: 3003-11 (2006) Article DOI: 10.1021/jm060047g BindingDB Entry DOI: 10.7270/Q25X28JQ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Tyrosine-protein phosphatase non-receptor type 1 (Homo sapiens (Human)) | BDBM50119685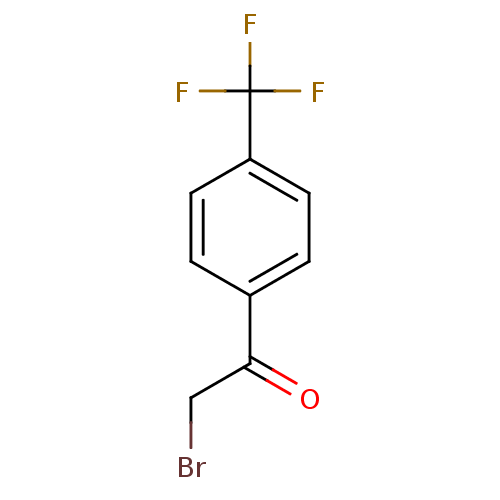 (2-Bromo-1-(4-trifluoromethyl-phenyl)-ethanone | CH...) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid UniChem Patents Similars | PubMed | 1.40E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Dissociation constant of the compound towards Protein-tyrosine phosphatase 1B receptor-inhibitor complex was determined using PNP as substrate | Bioorg Med Chem Lett 12: 3047-50 (2002) BindingDB Entry DOI: 10.7270/Q2FT8KCX | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Tyrosine-protein phosphatase non-receptor type 1 (Homo sapiens (Human)) | BDBM7880 (2-bromo-1-(4-phenylphenyl)ethan-1-one | CHEMBL4130...) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid UniChem Patents Similars | PubMed | 1.40E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Dissociation constant of the compound towards Protein-tyrosine phosphatase 1B receptor-inhibitor complex was determined using PNP as substrate | Bioorg Med Chem Lett 12: 3047-50 (2002) BindingDB Entry DOI: 10.7270/Q2FT8KCX | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| S-ribosylhomocysteine lyase (Bacillus subtilis) | BDBM50186747 ((2S)-2-AMINO-4-[(2R,3S)-2,3-DIHYDROXY-3-N-HYDROXYC...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | 1.96E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibition of zinc substituted Bacillus subtilis LuxS | J Med Chem 49: 3003-11 (2006) Article DOI: 10.1021/jm060047g BindingDB Entry DOI: 10.7270/Q25X28JQ | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Tyrosine-protein phosphatase non-receptor type 1 (Homo sapiens (Human)) | BDBM50119682 (2-Bromo-1-naphthalen-2-yl-ethanone | CHEMBL101423) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid UniChem Patents Similars | PubMed | 2.50E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Dissociation constant of the compound towards Protein-tyrosine phosphatase 1B receptor-inhibitor complex was determined using PNP as substrate | Bioorg Med Chem Lett 12: 3047-50 (2002) BindingDB Entry DOI: 10.7270/Q2FT8KCX | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Tyrosine-protein phosphatase non-receptor type 6 (Homo sapiens (Human)) | BDBM50119679 (2-[(S)-2-(2-{2-[4-(2-Bromo-acetyl)-phenoxy]-acetyl...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD antibodypedia GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | 2.90E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Dissociation constant of the compound towards SHP 1 receptor-inhibitor complex was determined using PNP as substrate | Bioorg Med Chem Lett 12: 3047-50 (2002) BindingDB Entry DOI: 10.7270/Q2FT8KCX | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Methionine aminopeptidase 1 (Homo sapiens (Human)) | BDBM50137352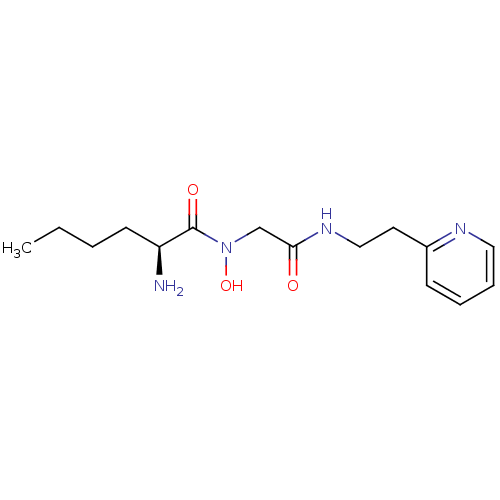 ((S)-2-Amino-hexanoic acid hydroxy-[(2-pyridin-2-yl...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem | PubMed | 3.30E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibitory activity of the compound against Methionine aminopeptidase 1 | Bioorg Med Chem Lett 14: 77-9 (2003) BindingDB Entry DOI: 10.7270/Q2DV1J90 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Peptide deformylase (Escherichia coli (strain K12)) | BDBM50291695 (CHEMBL84822 | H-PHOSPHONATE DERIVATIVE) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL KEGG PC cid PC sid UniChem Similars | PubMed | 3.70E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Ohio State University Curated by ChEMBL | Assay Description Compound was evaluated for inhibitory activity against Peptide deformylase | Bioorg Med Chem Lett 8: 2479-82 (1999) BindingDB Entry DOI: 10.7270/Q2HX1BTV | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| S-ribosylhomocysteine lyase (Bacillus subtilis) | BDBM50304164 (CHEMBL611818 | S-(5-Deoxy-3-O-methyl-D-ribofuranos...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 4.20E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Florida International University Curated by ChEMBL | Assay Description Inhibition of Bacillus subtilis LuxS | Bioorg Med Chem 17: 6699-706 (2009) Article DOI: 10.1016/j.bmc.2009.07.057 BindingDB Entry DOI: 10.7270/Q2PZ58XS | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (docked) | ||||||||||||
| S-ribosylhomocysteine lyase (Bacillus subtilis) | BDBM50304167 (CHEMBL610483 | S-(3,5-Dideoxy-3-fluoro-1-O-methyl-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 4.20E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Florida International University Curated by ChEMBL | Assay Description Inhibition of Bacillus subtilis LuxS | Bioorg Med Chem 17: 6699-706 (2009) Article DOI: 10.1016/j.bmc.2009.07.057 BindingDB Entry DOI: 10.7270/Q2PZ58XS | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Tyrosine-protein phosphatase non-receptor type 1 (Homo sapiens (Human)) | BDBM50119687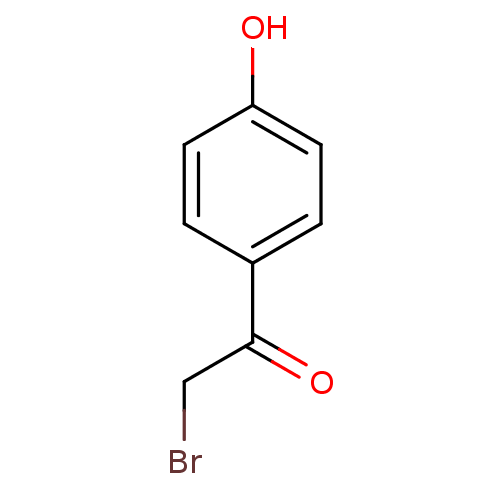 (2-Bromo-1-(4-hydroxy-phenyl)-ethanone | CHEMBL1029...) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid UniChem Patents Similars | PubMed | 4.30E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Dissociation constant of the compound towards Protein-tyrosine phosphatase 1B receptor-inhibitor complex was determined using PNP as substrate | Bioorg Med Chem Lett 12: 3047-50 (2002) BindingDB Entry DOI: 10.7270/Q2FT8KCX | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Displayed 1 to 50 (of 464 total ) | Next | Last >> |