

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5-hydroxytryptamine receptor 2C (Homo sapiens (Human)) | BDBM50161646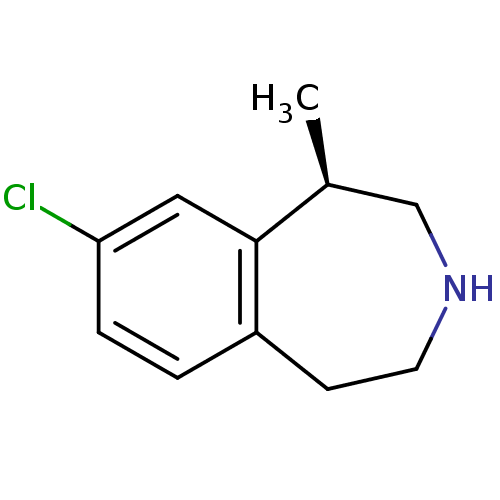 ((1R)-8-chloro-2,3,4,5-tetrahydro-1-methyl-1H-3-ben...) | PDB Reactome pathway KEGG UniProtKB/SwissProt DrugBank antibodypedia GoogleScholar AffyNet | Purchase CHEMBL PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 15 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Pfizer Inc. Curated by ChEMBL | Assay Description Displacement of [125I]DOI from human recombinant 5HT2C receptor expressed in HEK293 cells by scintillation counting | Bioorg Med Chem Lett 21: 2715-20 (2011) Article DOI: 10.1016/j.bmcl.2010.11.120 BindingDB Entry DOI: 10.7270/Q2HD7VZZ | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| 5-hydroxytryptamine receptor 2C (Homo sapiens (Human)) | BDBM50342539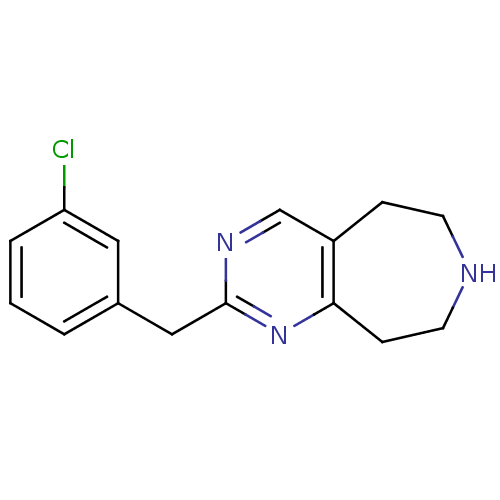 (2-(3-chlorobenzyl)-6,7,8,9-tetrahydro-5H-pyrimido[...) | PDB Reactome pathway KEGG UniProtKB/SwissProt DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents Similars | Article PubMed | 84 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Pfizer Inc. Curated by ChEMBL | Assay Description Displacement of [3H]meselurgine from human 5HT2C receptor expressed in Swiss mouse 3T3 cells by scintillation proximity assay | Bioorg Med Chem Lett 21: 2715-20 (2011) Article DOI: 10.1016/j.bmcl.2010.11.120 BindingDB Entry DOI: 10.7270/Q2HD7VZZ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| 5-hydroxytryptamine receptor 2C (Homo sapiens (Human)) | BDBM50342538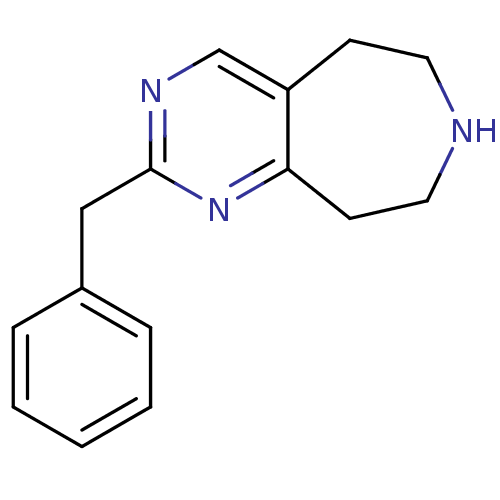 (2-benzyl-6,7,8,9-tetrahydro-5H-pyrimido[5,4-d]azep...) | PDB Reactome pathway KEGG UniProtKB/SwissProt DrugBank antibodypedia GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid UniChem Patents Similars | Article PubMed | 160 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Pfizer Inc. Curated by ChEMBL | Assay Description Displacement of [3H]meselurgine from human 5HT2C receptor expressed in Swiss mouse 3T3 cells by scintillation proximity assay | Bioorg Med Chem Lett 21: 2715-20 (2011) Article DOI: 10.1016/j.bmcl.2010.11.120 BindingDB Entry DOI: 10.7270/Q2HD7VZZ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| 5-hydroxytryptamine receptor 2C (Homo sapiens (Human)) | BDBM50161646 ((1R)-8-chloro-2,3,4,5-tetrahydro-1-methyl-1H-3-ben...) | PDB Reactome pathway KEGG UniProtKB/SwissProt DrugBank antibodypedia GoogleScholar AffyNet | Purchase CHEMBL PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 167 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Pfizer Inc. Curated by ChEMBL | Assay Description Displacement of [3H]meselurgine from human 5HT2C receptor expressed in Swiss mouse 3T3 cells by scintillation proximity assay | Bioorg Med Chem Lett 21: 2715-20 (2011) Article DOI: 10.1016/j.bmcl.2010.11.120 BindingDB Entry DOI: 10.7270/Q2HD7VZZ | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| 5-hydroxytryptamine receptor 2C (Homo sapiens (Human)) | BDBM50342547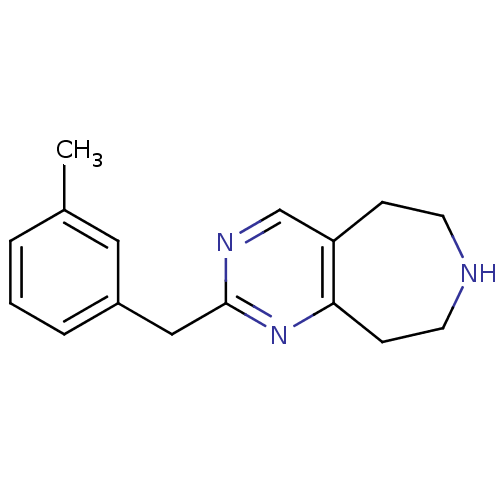 (2-(3-methylbenzyl)-6,7,8,9-tetrahydro-5H-pyrimido[...) | PDB Reactome pathway KEGG UniProtKB/SwissProt DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents Similars | Article PubMed | 244 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Pfizer Inc. Curated by ChEMBL | Assay Description Displacement of [3H]meselurgine from human 5HT2C receptor expressed in Swiss mouse 3T3 cells by scintillation proximity assay | Bioorg Med Chem Lett 21: 2715-20 (2011) Article DOI: 10.1016/j.bmcl.2010.11.120 BindingDB Entry DOI: 10.7270/Q2HD7VZZ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| 5-hydroxytryptamine receptor 2C (Homo sapiens (Human)) | BDBM50342548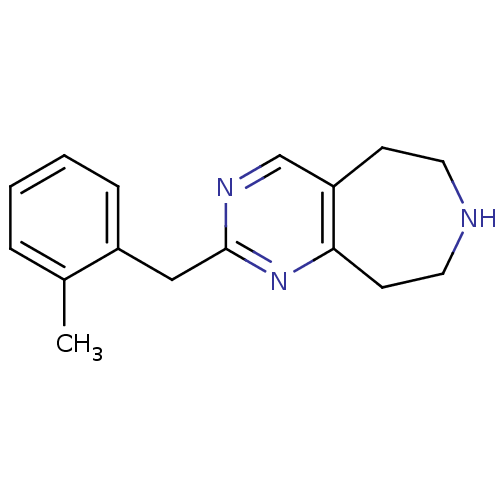 (2-(2-methylbenzyl)-6,7,8,9-tetrahydro-5H-pyrimido[...) | PDB Reactome pathway KEGG UniProtKB/SwissProt DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents Similars | Article PubMed | 305 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Pfizer Inc. Curated by ChEMBL | Assay Description Displacement of [3H]meselurgine from human 5HT2C receptor expressed in Swiss mouse 3T3 cells by scintillation proximity assay | Bioorg Med Chem Lett 21: 2715-20 (2011) Article DOI: 10.1016/j.bmcl.2010.11.120 BindingDB Entry DOI: 10.7270/Q2HD7VZZ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| 5-hydroxytryptamine receptor 2C (Homo sapiens (Human)) | BDBM50342544 (2-(3-(trifluoromethyl)benzyl)-6,7,8,9-tetrahydro-5...) | PDB Reactome pathway KEGG UniProtKB/SwissProt DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents Similars | Article PubMed | 308 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Pfizer Inc. Curated by ChEMBL | Assay Description Displacement of [3H]meselurgine from human 5HT2C receptor expressed in Swiss mouse 3T3 cells by scintillation proximity assay | Bioorg Med Chem Lett 21: 2715-20 (2011) Article DOI: 10.1016/j.bmcl.2010.11.120 BindingDB Entry DOI: 10.7270/Q2HD7VZZ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| 5-hydroxytryptamine receptor 2C (Homo sapiens (Human)) | BDBM50342549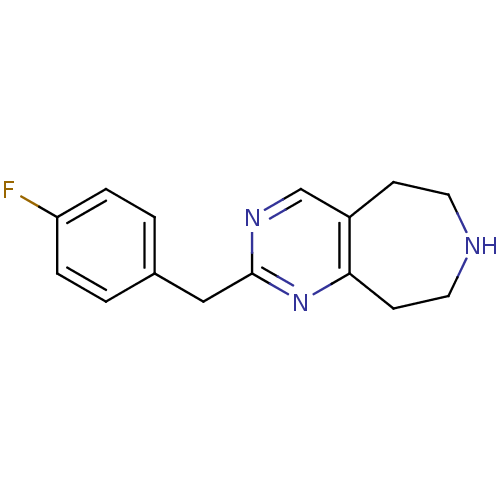 (2-(4-fluorobenzyl)-6,7,8,9-tetrahydro-5H-pyrimido[...) | PDB Reactome pathway KEGG UniProtKB/SwissProt DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents Similars | Article PubMed | 470 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Pfizer Inc. Curated by ChEMBL | Assay Description Displacement of [3H]meselurgine from human 5HT2C receptor expressed in Swiss mouse 3T3 cells by scintillation proximity assay | Bioorg Med Chem Lett 21: 2715-20 (2011) Article DOI: 10.1016/j.bmcl.2010.11.120 BindingDB Entry DOI: 10.7270/Q2HD7VZZ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| 5-hydroxytryptamine receptor 2C (Homo sapiens (Human)) | BDBM50342545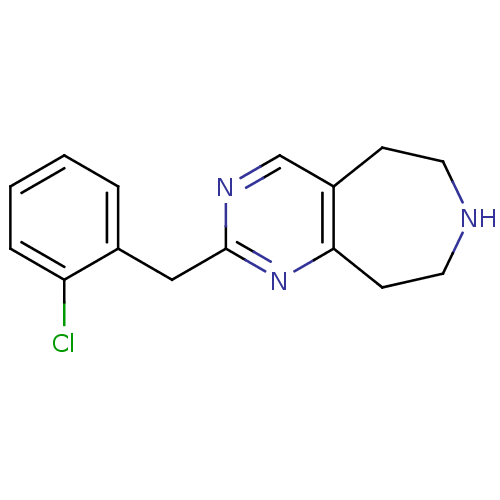 (2-(2-chlorobenzyl)-6,7,8,9-tetrahydro-5H-pyrimido[...) | PDB Reactome pathway KEGG UniProtKB/SwissProt DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 1.85E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Pfizer Inc. Curated by ChEMBL | Assay Description Displacement of [3H]meselurgine from human 5HT2C receptor expressed in Swiss mouse 3T3 cells by scintillation proximity assay | Bioorg Med Chem Lett 21: 2715-20 (2011) Article DOI: 10.1016/j.bmcl.2010.11.120 BindingDB Entry DOI: 10.7270/Q2HD7VZZ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Sliding-clamp-loader large/small subunit (Enterobacteria phage T4 (Bacteriophage T4)) | BDBM36319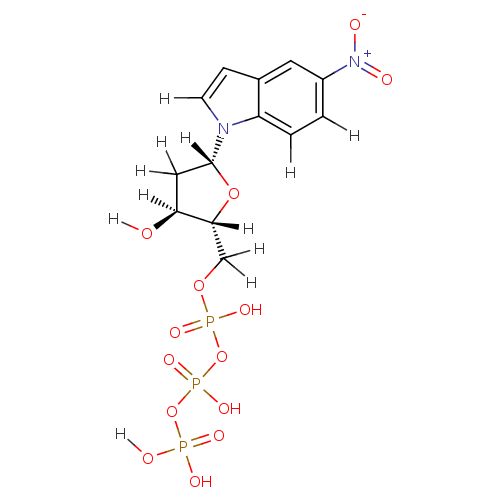 (5-nitro-indolyl- 2'-deoxyriboside triphosphate...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | KEGG MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | 4.80E+3 | -30.4 | n/a | n/a | n/a | n/a | n/a | 7.5 | 25 |
Case Western Reserve University | Assay Description ATPase assay contained 10 mM Mg2+, 1uM 13/20 DNA, 500uM (r)NTP or (d)NTP, 500 nM gp45, and 500 nM gp44/62 in T4 buffer. ATPase activity was measured ... | ACS Chem Biol 5: 183-94 (2010) Article DOI: 10.1021/cb900218c BindingDB Entry DOI: 10.7270/Q26M356K | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Complex of Sliding-clamp-loader large subunit [R175L] and Sliding-clamp-loader small subunit (bacteriophage T4) | BDBM36324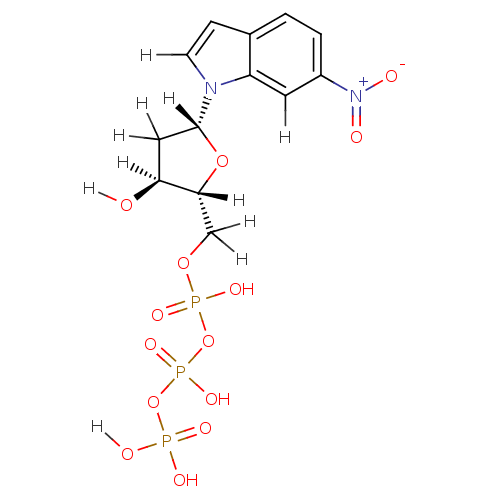 (6-nitro-indolyl- 2'-deoxyriboside triphosphate...) | PDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | KEGG PC cid PC sid UniChem Similars | Article PubMed | 5.00E+3 | -30.3 | n/a | n/a | n/a | n/a | n/a | 7.5 | 25 |
Case Western Reserve University | Assay Description ATPase assay contained 10 mM Mg2+, 1uM 13/20 DNA, 500uM (r)NTP or (d)NTP, 500 nM gp45, and 500 nM gp44/62 in T4 buffer. ATPase activity was measured ... | ACS Chem Biol 5: 183-94 (2010) Article DOI: 10.1021/cb900218c BindingDB Entry DOI: 10.7270/Q26M356K | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Sliding-clamp-loader large/small subunit (Enterobacteria phage T4 (Bacteriophage T4)) | BDBM36324 (6-nitro-indolyl- 2'-deoxyriboside triphosphate...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | KEGG PC cid PC sid UniChem Similars | Article PubMed | 5.10E+3 | -30.2 | n/a | n/a | n/a | n/a | n/a | 7.5 | 25 |
Case Western Reserve University | Assay Description ATPase assay contained 10 mM Mg2+, 1uM 13/20 DNA, 500uM (r)NTP or (d)NTP, 500 nM gp45, and 500 nM gp44/62 in T4 buffer. ATPase activity was measured ... | ACS Chem Biol 5: 183-94 (2010) Article DOI: 10.1021/cb900218c BindingDB Entry DOI: 10.7270/Q26M356K | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA polymerase III subunit tau (Escherichia coli) | BDBM36322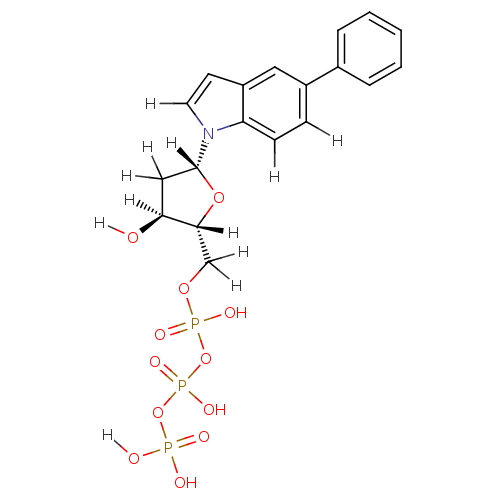 (5-phenyl-indolyl- 2'-deoxyriboside triphosphat...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | KEGG PC cid PC sid UniChem Similars | Article PubMed | 7.50E+3 | -30.4 | n/a | n/a | n/a | n/a | n/a | 7.5 | 37 |
Case Western Reserve University | Assay Description ATPase assay contained 10 mM Mg2+, 1uM 13/20 DNA, 500uM (r)NTP or (d)NTP, 500 nM gp45, and 500 nM gp44/62 in T4 buffer. ATPase activity was measured ... | ACS Chem Biol 5: 183-94 (2010) Article DOI: 10.1021/cb900218c BindingDB Entry DOI: 10.7270/Q26M356K | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA polymerase III subunit tau (Escherichia coli) | BDBM36324 (6-nitro-indolyl- 2'-deoxyriboside triphosphate...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | KEGG PC cid PC sid UniChem Similars | Article PubMed | 8.10E+3 | -30.2 | n/a | n/a | n/a | n/a | n/a | 7.5 | 37 |
Case Western Reserve University | Assay Description ATPase assay contained 10 mM Mg2+, 1uM 13/20 DNA, 500uM (r)NTP or (d)NTP, 500 nM gp45, and 500 nM gp44/62 in T4 buffer. ATPase activity was measured ... | ACS Chem Biol 5: 183-94 (2010) Article DOI: 10.1021/cb900218c BindingDB Entry DOI: 10.7270/Q26M356K | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA polymerase III subunit tau (Escherichia coli) | BDBM36316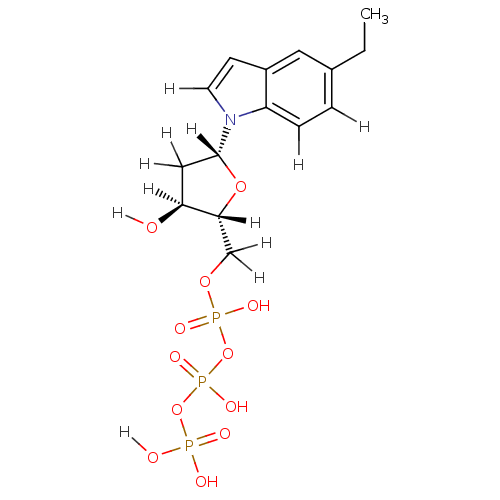 (5-ethyl-indolyl- 2'-deoxyriboside triphosphate...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | KEGG PC cid PC sid UniChem Similars | Article PubMed | 9.00E+3 | -30.0 | n/a | n/a | n/a | n/a | n/a | 7.5 | 37 |
Case Western Reserve University | Assay Description ATPase assay contained 10 mM Mg2+, 1uM 13/20 DNA, 500uM (r)NTP or (d)NTP, 500 nM gp45, and 500 nM gp44/62 in T4 buffer. ATPase activity was measured ... | ACS Chem Biol 5: 183-94 (2010) Article DOI: 10.1021/cb900218c BindingDB Entry DOI: 10.7270/Q26M356K | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Complex of Sliding-clamp-loader large subunit [R175K] and Sliding-clamp-loader small subunit (bacteriophage T4) | BDBM36324 (6-nitro-indolyl- 2'-deoxyriboside triphosphate...) | PDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | KEGG PC cid PC sid UniChem Similars | Article PubMed | 9.10E+3 | -28.8 | n/a | n/a | n/a | n/a | n/a | 7.5 | 25 |
Case Western Reserve University | Assay Description ATPase assay contained 10 mM Mg2+, 1uM 13/20 DNA, 500uM (r)NTP or (d)NTP, 500 nM gp45, and 500 nM gp44/62 in T4 buffer. ATPase activity was measured ... | ACS Chem Biol 5: 183-94 (2010) Article DOI: 10.1021/cb900218c BindingDB Entry DOI: 10.7270/Q26M356K | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Sliding-clamp-loader large/small subunit (Enterobacteria phage T4 (Bacteriophage T4)) | BDBM36321 (5-cyclohexene-indolyl- 2'-deoxyriboside tripho...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | KEGG PC cid PC sid UniChem Similars | Article PubMed | 1.00E+4 | -28.5 | n/a | n/a | n/a | n/a | n/a | 7.5 | 25 |
Case Western Reserve University | Assay Description ATPase assay contained 10 mM Mg2+, 1uM 13/20 DNA, 500uM (r)NTP or (d)NTP, 500 nM gp45, and 500 nM gp44/62 in T4 buffer. ATPase activity was measured ... | ACS Chem Biol 5: 183-94 (2010) Article DOI: 10.1021/cb900218c BindingDB Entry DOI: 10.7270/Q26M356K | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Complex of Sliding-clamp-loader large subunit [R175L] and Sliding-clamp-loader small subunit (bacteriophage T4) | BDBM36316 (5-ethyl-indolyl- 2'-deoxyriboside triphosphate...) | PDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | KEGG PC cid PC sid UniChem Similars | Article PubMed | 1.10E+4 | -28.3 | n/a | n/a | n/a | n/a | n/a | 7.5 | 25 |
Case Western Reserve University | Assay Description ATPase assay contained 10 mM Mg2+, 1uM 13/20 DNA, 500uM (r)NTP or (d)NTP, 500 nM gp45, and 500 nM gp44/62 in T4 buffer. ATPase activity was measured ... | ACS Chem Biol 5: 183-94 (2010) Article DOI: 10.1021/cb900218c BindingDB Entry DOI: 10.7270/Q26M356K | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA polymerase III subunit tau (Escherichia coli) | BDBM36323 (4-nitro-indolyl- 2'-deoxyriboside triphosphate...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | KEGG PC cid PC sid UniChem Similars | Article PubMed | 1.11E+4 | -29.4 | n/a | n/a | n/a | n/a | n/a | 7.5 | 37 |
Case Western Reserve University | Assay Description ATPase assay contained 10 mM Mg2+, 1uM 13/20 DNA, 500uM (r)NTP or (d)NTP, 500 nM gp45, and 500 nM gp44/62 in T4 buffer. ATPase activity was measured ... | ACS Chem Biol 5: 183-94 (2010) Article DOI: 10.1021/cb900218c BindingDB Entry DOI: 10.7270/Q26M356K | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA polymerase III subunit tau (Escherichia coli) | BDBM36321 (5-cyclohexene-indolyl- 2'-deoxyriboside tripho...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | KEGG PC cid PC sid UniChem Similars | Article PubMed | 1.18E+4 | -29.3 | n/a | n/a | n/a | n/a | n/a | 7.5 | 37 |
Case Western Reserve University | Assay Description ATPase assay contained 10 mM Mg2+, 1uM 13/20 DNA, 500uM (r)NTP or (d)NTP, 500 nM gp45, and 500 nM gp44/62 in T4 buffer. ATPase activity was measured ... | ACS Chem Biol 5: 183-94 (2010) Article DOI: 10.1021/cb900218c BindingDB Entry DOI: 10.7270/Q26M356K | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA polymerase III subunit tau (Escherichia coli) | BDBM36312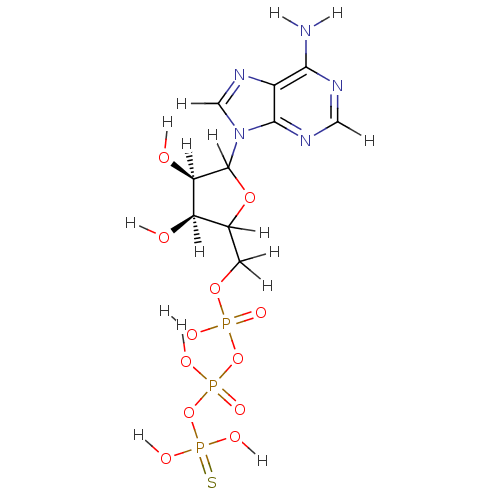 (ATP{gamma}S | adenosine 5'-O-(3-thiotriphospha...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase PC cid PC sid UniChem Similars | Article PubMed | 1.30E+4 | -29.0 | n/a | n/a | n/a | n/a | n/a | 7.5 | 37 |
Case Western Reserve University | Assay Description ATPase assay contained 10 mM Mg2+, 1uM 13/20 DNA, 500uM (r)NTP or (d)NTP, 500 nM gp45, and 500 nM gp44/62 in T4 buffer. ATPase activity was measured ... | ACS Chem Biol 5: 183-94 (2010) Article DOI: 10.1021/cb900218c BindingDB Entry DOI: 10.7270/Q26M356K | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Complex of Sliding-clamp-loader large subunit [R175K] and Sliding-clamp-loader small subunit (bacteriophage T4) | BDBM36319 (5-nitro-indolyl- 2'-deoxyriboside triphosphate...) | PDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | KEGG MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | 1.38E+4 | -27.7 | n/a | n/a | n/a | n/a | n/a | 7.5 | 25 |
Case Western Reserve University | Assay Description ATPase assay contained 10 mM Mg2+, 1uM 13/20 DNA, 500uM (r)NTP or (d)NTP, 500 nM gp45, and 500 nM gp44/62 in T4 buffer. ATPase activity was measured ... | ACS Chem Biol 5: 183-94 (2010) Article DOI: 10.1021/cb900218c BindingDB Entry DOI: 10.7270/Q26M356K | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Complex of Sliding-clamp-loader large subunit [R175K] and Sliding-clamp-loader small subunit (bacteriophage T4) | BDBM36312 (ATP{gamma}S | adenosine 5'-O-(3-thiotriphospha...) | PDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase PC cid PC sid UniChem Similars | Article PubMed | 1.40E+4 | -27.7 | n/a | n/a | n/a | n/a | n/a | 7.5 | 25 |
Case Western Reserve University | Assay Description ATPase assay contained 10 mM Mg2+, 1uM 13/20 DNA, 500uM (r)NTP or (d)NTP, 500 nM gp45, and 500 nM gp44/62 in T4 buffer. ATPase activity was measured ... | ACS Chem Biol 5: 183-94 (2010) Article DOI: 10.1021/cb900218c BindingDB Entry DOI: 10.7270/Q26M356K | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Complex of Sliding-clamp-loader large subunit [R175L] and Sliding-clamp-loader small subunit (bacteriophage T4) | BDBM36319 (5-nitro-indolyl- 2'-deoxyriboside triphosphate...) | PDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | KEGG MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | 1.64E+4 | -27.3 | n/a | n/a | n/a | n/a | n/a | 7.5 | 25 |
Case Western Reserve University | Assay Description ATPase assay contained 10 mM Mg2+, 1uM 13/20 DNA, 500uM (r)NTP or (d)NTP, 500 nM gp45, and 500 nM gp44/62 in T4 buffer. ATPase activity was measured ... | ACS Chem Biol 5: 183-94 (2010) Article DOI: 10.1021/cb900218c BindingDB Entry DOI: 10.7270/Q26M356K | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Angiotensin-converting enzyme (Oryctolagus cuniculus) | BDBM50559562 (CHEMBL4762834) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 1.80E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA | Assay Description Inhibition of rabbit ACE using Abz-FRK(DNP)-P as substrate measured for every 30 sec for 5 mins by UV-fluorescence based assay | Citation and Details Article DOI: 10.1021/acsmedchemlett.0c00554 BindingDB Entry DOI: 10.7270/Q2X63RNT | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA polymerase III subunit tau (Escherichia coli) | BDBM36319 (5-nitro-indolyl- 2'-deoxyriboside triphosphate...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | KEGG MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | 2.17E+4 | -27.7 | n/a | n/a | n/a | n/a | n/a | 7.5 | 37 |
Case Western Reserve University | Assay Description ATPase assay contained 10 mM Mg2+, 1uM 13/20 DNA, 500uM (r)NTP or (d)NTP, 500 nM gp45, and 500 nM gp44/62 in T4 buffer. ATPase activity was measured ... | ACS Chem Biol 5: 183-94 (2010) Article DOI: 10.1021/cb900218c BindingDB Entry DOI: 10.7270/Q26M356K | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA polymerase III subunit tau (Escherichia coli) | BDBM36320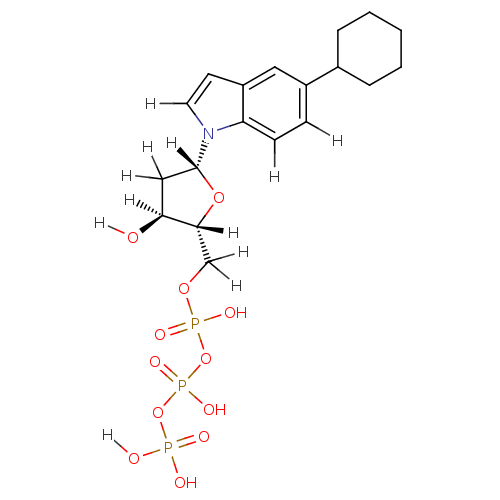 (5-cyclohexyl-indolyl- 2'-deoxyriboside triphos...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | KEGG PC cid PC sid UniChem Similars | Article PubMed | 2.40E+4 | -27.4 | n/a | n/a | n/a | n/a | n/a | 7.5 | 37 |
Case Western Reserve University | Assay Description ATPase assay contained 10 mM Mg2+, 1uM 13/20 DNA, 500uM (r)NTP or (d)NTP, 500 nM gp45, and 500 nM gp44/62 in T4 buffer. ATPase activity was measured ... | ACS Chem Biol 5: 183-94 (2010) Article DOI: 10.1021/cb900218c BindingDB Entry DOI: 10.7270/Q26M356K | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Complex of Sliding-clamp-loader large subunit [R175K] and Sliding-clamp-loader small subunit (bacteriophage T4) | BDBM36316 (5-ethyl-indolyl- 2'-deoxyriboside triphosphate...) | PDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | KEGG PC cid PC sid UniChem Similars | Article PubMed | 2.52E+4 | -26.2 | n/a | n/a | n/a | n/a | n/a | 7.5 | 25 |
Case Western Reserve University | Assay Description ATPase assay contained 10 mM Mg2+, 1uM 13/20 DNA, 500uM (r)NTP or (d)NTP, 500 nM gp45, and 500 nM gp44/62 in T4 buffer. ATPase activity was measured ... | ACS Chem Biol 5: 183-94 (2010) Article DOI: 10.1021/cb900218c BindingDB Entry DOI: 10.7270/Q26M356K | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Complex of Sliding-clamp-loader large subunit [R175K] and Sliding-clamp-loader small subunit (bacteriophage T4) | BDBM36317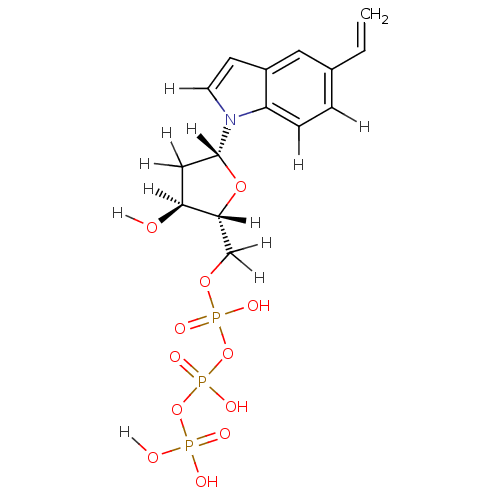 (5-ethylene-indolyl- 2'-deoxyriboside triphosph...) | PDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | KEGG PC cid PC sid UniChem Similars | Article PubMed | 2.62E+4 | -26.2 | n/a | n/a | n/a | n/a | n/a | 7.5 | 25 |
Case Western Reserve University | Assay Description ATPase assay contained 10 mM Mg2+, 1uM 13/20 DNA, 500uM (r)NTP or (d)NTP, 500 nM gp45, and 500 nM gp44/62 in T4 buffer. ATPase activity was measured ... | ACS Chem Biol 5: 183-94 (2010) Article DOI: 10.1021/cb900218c BindingDB Entry DOI: 10.7270/Q26M356K | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Sliding-clamp-loader large/small subunit (Enterobacteria phage T4 (Bacteriophage T4)) | BDBM36312 (ATP{gamma}S | adenosine 5'-O-(3-thiotriphospha...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | Purchase PC cid PC sid UniChem Similars | Article PubMed | 2.89E+4 | -25.9 | n/a | n/a | n/a | n/a | n/a | 7.5 | 25 |
Case Western Reserve University | Assay Description ATPase assay contained 10 mM Mg2+, 1uM 13/20 DNA, 500uM (r)NTP or (d)NTP, 500 nM gp45, and 500 nM gp44/62 in T4 buffer. ATPase activity was measured ... | ACS Chem Biol 5: 183-94 (2010) Article DOI: 10.1021/cb900218c BindingDB Entry DOI: 10.7270/Q26M356K | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA polymerase III subunit tau (Escherichia coli) | BDBM36317 (5-ethylene-indolyl- 2'-deoxyriboside triphosph...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | KEGG PC cid PC sid UniChem Similars | Article PubMed | 3.00E+4 | -26.9 | n/a | n/a | n/a | n/a | n/a | 7.5 | 37 |
Case Western Reserve University | Assay Description ATPase assay contained 10 mM Mg2+, 1uM 13/20 DNA, 500uM (r)NTP or (d)NTP, 500 nM gp45, and 500 nM gp44/62 in T4 buffer. ATPase activity was measured ... | ACS Chem Biol 5: 183-94 (2010) Article DOI: 10.1021/cb900218c BindingDB Entry DOI: 10.7270/Q26M356K | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Complex of Sliding-clamp-loader large subunit [R175K] and Sliding-clamp-loader small subunit (bacteriophage T4) | BDBM36323 (4-nitro-indolyl- 2'-deoxyriboside triphosphate...) | PDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | KEGG PC cid PC sid UniChem Similars | Article PubMed | 3.02E+4 | -25.8 | n/a | n/a | n/a | n/a | n/a | 7.5 | 25 |
Case Western Reserve University | Assay Description ATPase assay contained 10 mM Mg2+, 1uM 13/20 DNA, 500uM (r)NTP or (d)NTP, 500 nM gp45, and 500 nM gp44/62 in T4 buffer. ATPase activity was measured ... | ACS Chem Biol 5: 183-94 (2010) Article DOI: 10.1021/cb900218c BindingDB Entry DOI: 10.7270/Q26M356K | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Angiotensin-converting enzyme (Oryctolagus cuniculus) | BDBM50559561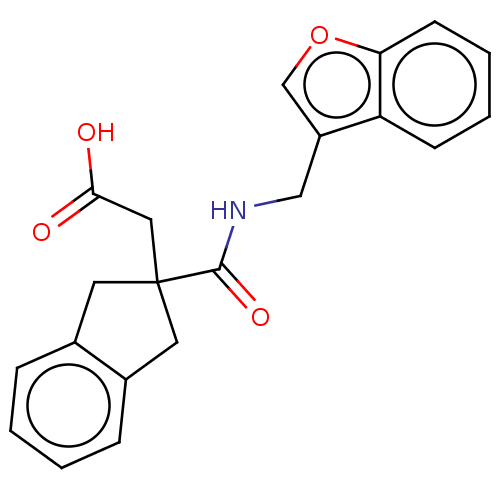 (CHEMBL4760006) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 3.13E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA | Assay Description Inhibition of rabbit ACE using Abz-FRK(DNP)-P as substrate measured for every 30 sec for 5 mins by UV-fluorescence based assay | Citation and Details Article DOI: 10.1021/acsmedchemlett.0c00554 BindingDB Entry DOI: 10.7270/Q2X63RNT | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Complex of Sliding-clamp-loader large subunit [R175L] and Sliding-clamp-loader small subunit (bacteriophage T4) | BDBM36312 (ATP{gamma}S | adenosine 5'-O-(3-thiotriphospha...) | PDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase PC cid PC sid UniChem Similars | Article PubMed | 3.32E+4 | -25.6 | n/a | n/a | n/a | n/a | n/a | 7.5 | 25 |
Case Western Reserve University | Assay Description ATPase assay contained 10 mM Mg2+, 1uM 13/20 DNA, 500uM (r)NTP or (d)NTP, 500 nM gp45, and 500 nM gp44/62 in T4 buffer. ATPase activity was measured ... | ACS Chem Biol 5: 183-94 (2010) Article DOI: 10.1021/cb900218c BindingDB Entry DOI: 10.7270/Q26M356K | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Complex of Sliding-clamp-loader large subunit [R175L] and Sliding-clamp-loader small subunit (bacteriophage T4) | BDBM36317 (5-ethylene-indolyl- 2'-deoxyriboside triphosph...) | PDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | KEGG PC cid PC sid UniChem Similars | Article PubMed | 3.40E+4 | -25.5 | n/a | n/a | n/a | n/a | n/a | 7.5 | 25 |
Case Western Reserve University | Assay Description ATPase assay contained 10 mM Mg2+, 1uM 13/20 DNA, 500uM (r)NTP or (d)NTP, 500 nM gp45, and 500 nM gp44/62 in T4 buffer. ATPase activity was measured ... | ACS Chem Biol 5: 183-94 (2010) Article DOI: 10.1021/cb900218c BindingDB Entry DOI: 10.7270/Q26M356K | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA polymerase III subunit tau (Escherichia coli) | BDBM36315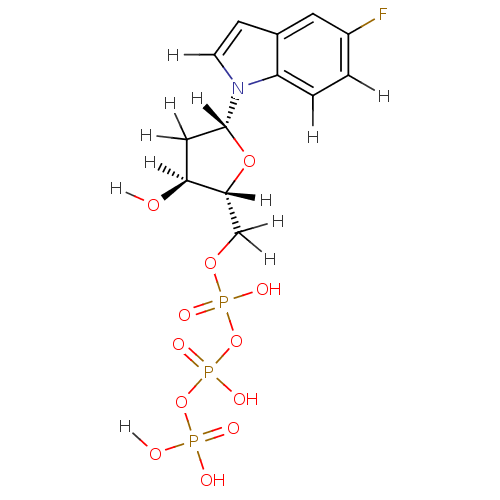 (5-fluoro-indolyl- 2'-deoxyriboside triphosphat...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | KEGG PC cid PC sid UniChem Similars | Article PubMed | 3.40E+4 | -26.5 | n/a | n/a | n/a | n/a | n/a | 7.5 | 37 |
Case Western Reserve University | Assay Description ATPase assay contained 10 mM Mg2+, 1uM 13/20 DNA, 500uM (r)NTP or (d)NTP, 500 nM gp45, and 500 nM gp44/62 in T4 buffer. ATPase activity was measured ... | ACS Chem Biol 5: 183-94 (2010) Article DOI: 10.1021/cb900218c BindingDB Entry DOI: 10.7270/Q26M356K | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Sliding-clamp-loader large/small subunit (Enterobacteria phage T4 (Bacteriophage T4)) | BDBM36323 (4-nitro-indolyl- 2'-deoxyriboside triphosphate...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | KEGG PC cid PC sid UniChem Similars | Article PubMed | 3.42E+4 | -25.5 | n/a | n/a | n/a | n/a | n/a | 7.5 | 25 |
Case Western Reserve University | Assay Description ATPase assay contained 10 mM Mg2+, 1uM 13/20 DNA, 500uM (r)NTP or (d)NTP, 500 nM gp45, and 500 nM gp44/62 in T4 buffer. ATPase activity was measured ... | ACS Chem Biol 5: 183-94 (2010) Article DOI: 10.1021/cb900218c BindingDB Entry DOI: 10.7270/Q26M356K | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Sliding-clamp-loader large/small subunit (Enterobacteria phage T4 (Bacteriophage T4)) | BDBM36318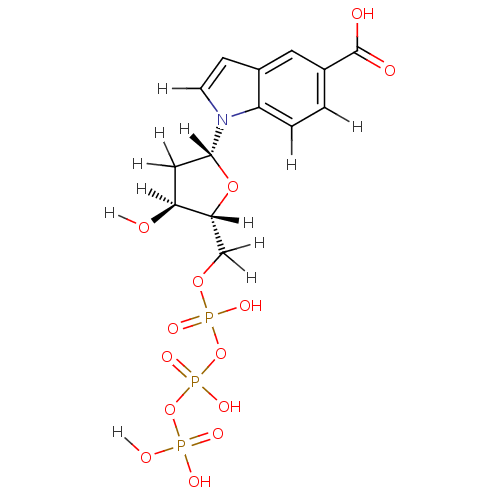 (5-carboxylate-indolyl- 2'-deoxyriboside tripho...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | KEGG PC cid PC sid UniChem Similars | Article PubMed | 3.70E+4 | -25.3 | n/a | n/a | n/a | n/a | n/a | 7.5 | 25 |
Case Western Reserve University | Assay Description ATPase assay contained 10 mM Mg2+, 1uM 13/20 DNA, 500uM (r)NTP or (d)NTP, 500 nM gp45, and 500 nM gp44/62 in T4 buffer. ATPase activity was measured ... | ACS Chem Biol 5: 183-94 (2010) Article DOI: 10.1021/cb900218c BindingDB Entry DOI: 10.7270/Q26M356K | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA polymerase III subunit tau (Escherichia coli) | BDBM36318 (5-carboxylate-indolyl- 2'-deoxyriboside tripho...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | KEGG PC cid PC sid UniChem Similars | Article PubMed | 4.00E+4 | -26.1 | n/a | n/a | n/a | n/a | n/a | 7.5 | 37 |
Case Western Reserve University | Assay Description ATPase assay contained 10 mM Mg2+, 1uM 13/20 DNA, 500uM (r)NTP or (d)NTP, 500 nM gp45, and 500 nM gp44/62 in T4 buffer. ATPase activity was measured ... | ACS Chem Biol 5: 183-94 (2010) Article DOI: 10.1021/cb900218c BindingDB Entry DOI: 10.7270/Q26M356K | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Complex of Sliding-clamp-loader large subunit [R175K] and Sliding-clamp-loader small subunit (bacteriophage T4) | BDBM36318 (5-carboxylate-indolyl- 2'-deoxyriboside tripho...) | PDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | KEGG PC cid PC sid UniChem Similars | Article PubMed | 4.06E+4 | -25.1 | n/a | n/a | n/a | n/a | n/a | 7.5 | 25 |
Case Western Reserve University | Assay Description ATPase assay contained 10 mM Mg2+, 1uM 13/20 DNA, 500uM (r)NTP or (d)NTP, 500 nM gp45, and 500 nM gp44/62 in T4 buffer. ATPase activity was measured ... | ACS Chem Biol 5: 183-94 (2010) Article DOI: 10.1021/cb900218c BindingDB Entry DOI: 10.7270/Q26M356K | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Angiotensin-converting enzyme (Oryctolagus cuniculus) | BDBM50559558 (CHEMBL4764936) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 4.06E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA | Assay Description Inhibition of rabbit ACE using Abz-FRK(DNP)-P as substrate measured for every 30 sec for 5 mins by UV-fluorescence based assay | Citation and Details Article DOI: 10.1021/acsmedchemlett.0c00554 BindingDB Entry DOI: 10.7270/Q2X63RNT | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Complex of Sliding-clamp-loader large subunit [R175L] and Sliding-clamp-loader small subunit (bacteriophage T4) | BDBM36323 (4-nitro-indolyl- 2'-deoxyriboside triphosphate...) | PDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | KEGG PC cid PC sid UniChem Similars | Article PubMed | 4.16E+4 | -25.0 | n/a | n/a | n/a | n/a | n/a | 7.5 | 25 |
Case Western Reserve University | Assay Description ATPase assay contained 10 mM Mg2+, 1uM 13/20 DNA, 500uM (r)NTP or (d)NTP, 500 nM gp45, and 500 nM gp44/62 in T4 buffer. ATPase activity was measured ... | ACS Chem Biol 5: 183-94 (2010) Article DOI: 10.1021/cb900218c BindingDB Entry DOI: 10.7270/Q26M356K | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Sliding-clamp-loader large/small subunit (Enterobacteria phage T4 (Bacteriophage T4)) | BDBM36322 (5-phenyl-indolyl- 2'-deoxyriboside triphosphat...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | KEGG PC cid PC sid UniChem Similars | Article PubMed | 4.20E+4 | -25.0 | n/a | n/a | n/a | n/a | n/a | 7.5 | 25 |
Case Western Reserve University | Assay Description ATPase assay contained 10 mM Mg2+, 1uM 13/20 DNA, 500uM (r)NTP or (d)NTP, 500 nM gp45, and 500 nM gp44/62 in T4 buffer. ATPase activity was measured ... | ACS Chem Biol 5: 183-94 (2010) Article DOI: 10.1021/cb900218c BindingDB Entry DOI: 10.7270/Q26M356K | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA polymerase III subunit tau (Escherichia coli) | BDBM36314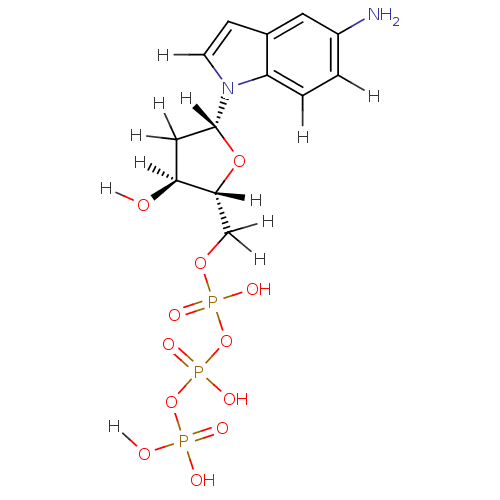 (5-amino-indolyl- 2'-deoxyriboside triphosphate...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | KEGG PC cid PC sid UniChem Similars | Article PubMed | 4.50E+4 | -25.8 | n/a | n/a | n/a | n/a | n/a | 7.5 | 37 |
Case Western Reserve University | Assay Description ATPase assay contained 10 mM Mg2+, 1uM 13/20 DNA, 500uM (r)NTP or (d)NTP, 500 nM gp45, and 500 nM gp44/62 in T4 buffer. ATPase activity was measured ... | ACS Chem Biol 5: 183-94 (2010) Article DOI: 10.1021/cb900218c BindingDB Entry DOI: 10.7270/Q26M356K | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Sliding-clamp-loader large/small subunit (Enterobacteria phage T4 (Bacteriophage T4)) | BDBM36320 (5-cyclohexyl-indolyl- 2'-deoxyriboside triphos...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | KEGG PC cid PC sid UniChem Similars | Article PubMed | 4.75E+4 | -24.7 | n/a | n/a | n/a | n/a | n/a | 7.5 | 25 |
Case Western Reserve University | Assay Description ATPase assay contained 10 mM Mg2+, 1uM 13/20 DNA, 500uM (r)NTP or (d)NTP, 500 nM gp45, and 500 nM gp44/62 in T4 buffer. ATPase activity was measured ... | ACS Chem Biol 5: 183-94 (2010) Article DOI: 10.1021/cb900218c BindingDB Entry DOI: 10.7270/Q26M356K | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Angiotensin-converting enzyme (Oryctolagus cuniculus) | BDBM50559566 (CHEMBL4749721) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | >5.70E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA | Assay Description Inhibition of rabbit ACE using Abz-FRK(DNP)-P as substrate measured for every 30 sec for 5 mins by UV-fluorescence based assay | Citation and Details Article DOI: 10.1021/acsmedchemlett.0c00554 BindingDB Entry DOI: 10.7270/Q2X63RNT | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Angiotensin-converting enzyme (Oryctolagus cuniculus) | BDBM50559567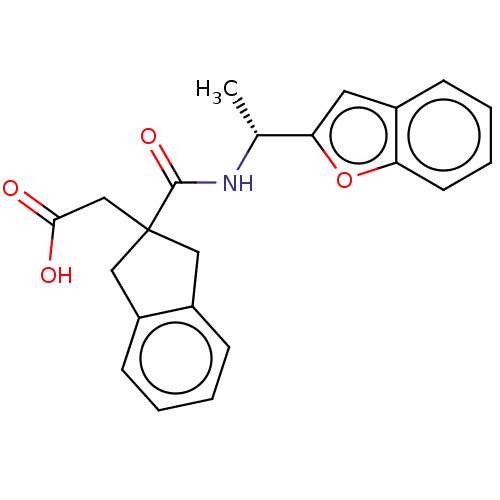 (CHEMBL4760989) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | >5.70E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA | Assay Description Inhibition of rabbit ACE using Abz-FRK(DNP)-P as substrate measured for every 30 sec for 5 mins by UV-fluorescence based assay | Citation and Details Article DOI: 10.1021/acsmedchemlett.0c00554 BindingDB Entry DOI: 10.7270/Q2X63RNT | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Angiotensin-converting enzyme (Oryctolagus cuniculus) | BDBM50559568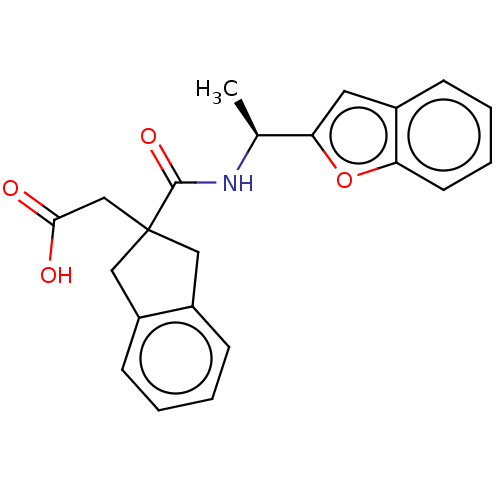 (CHEMBL4748733) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | >5.70E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA | Assay Description Inhibition of rabbit ACE using Abz-FRK(DNP)-P as substrate measured for every 30 sec for 5 mins by UV-fluorescence based assay | Citation and Details Article DOI: 10.1021/acsmedchemlett.0c00554 BindingDB Entry DOI: 10.7270/Q2X63RNT | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Angiotensin-converting enzyme (Oryctolagus cuniculus) | BDBM50559569 (CHEMBL4761199) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | >5.70E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA | Assay Description Inhibition of rabbit ACE using Abz-FRK(DNP)-P as substrate measured for every 30 sec for 5 mins by UV-fluorescence based assay | Citation and Details Article DOI: 10.1021/acsmedchemlett.0c00554 BindingDB Entry DOI: 10.7270/Q2X63RNT | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Angiotensin-converting enzyme (Oryctolagus cuniculus) | BDBM50559570 (CHEMBL4760832) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | >5.70E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA | Assay Description Inhibition of rabbit ACE using Abz-FRK(DNP)-P as substrate measured for every 30 sec for 5 mins by UV-fluorescence based assay | Citation and Details Article DOI: 10.1021/acsmedchemlett.0c00554 BindingDB Entry DOI: 10.7270/Q2X63RNT | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Displayed 1 to 50 (of 1946 total ) | Next | Last >> |