

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DNA gyrase subunit B (Escherichia coli) | BDBM50330317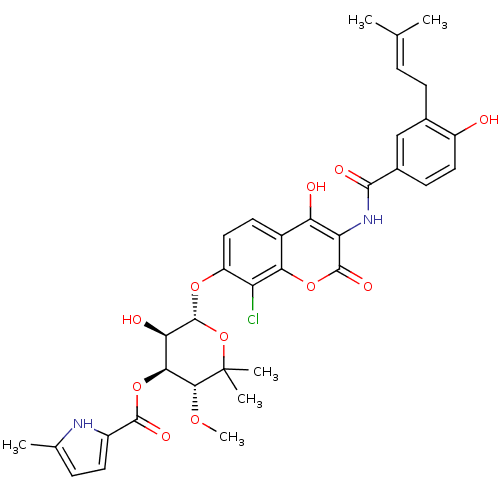 (5-Methyl-1H-pyrrole-2-carboxylic acid (3R,4S,5R,6S...) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | Purchase CHEMBL MCE KEGG MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | n/a | n/a | n/a | 1.20 | n/a | n/a | n/a | n/a | n/a |
University of Ljubljana Curated by ChEMBL | Assay Description Binding affinity to Escherichia coli GyrB | J Med Chem 54: 915-29 (2011) Article DOI: 10.1021/jm101121s BindingDB Entry DOI: 10.7270/Q2NS0W23 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50226181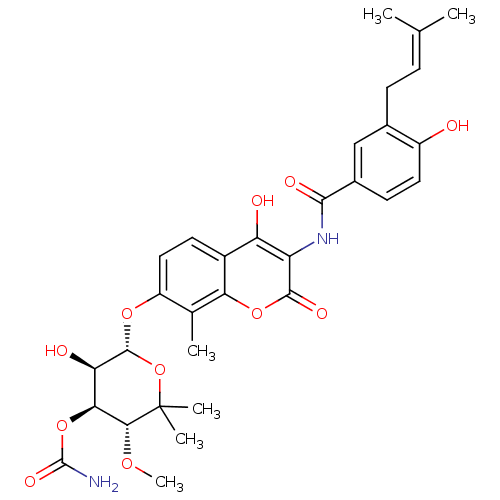 ((3R,4S,5R,6R)-5-hydroxy-6-(4-hydroxy-3-(4-hydroxy-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE KEGG MMDB PC cid PC sid PDB UniChem Similars | MMDB Article PubMed | n/a | n/a | n/a | 7 | n/a | n/a | n/a | n/a | n/a |
R. C. Patel Institute of Pharmaceutical Education and Research Curated by ChEMBL | Assay Description Binding affinity to Escherichia coli H560 DNA gyrase B | Eur J Med Chem 124: 160-185 (2016) Article DOI: 10.1016/j.ejmech.2016.08.034 BindingDB Entry DOI: 10.7270/Q2K64M38 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit A/B (Escherichia coli (strain K12)) | BDBM50095149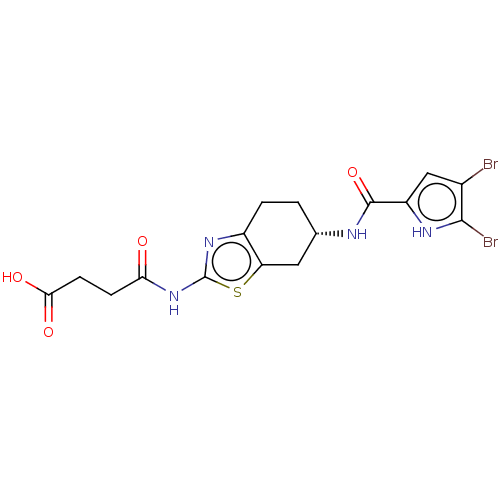 (CHEMBL3589536) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents Similars | Article PubMed | n/a | n/a | n/a | 10 | n/a | n/a | n/a | n/a | n/a |
University of Ljubljana Curated by ChEMBL | Assay Description Binding affinity to Escherichia coli DNA gyrase subunit B N-terminal 24 kDa domain by SPR assay | J Med Chem 58: 5501-21 (2015) Article DOI: 10.1021/acs.jmedchem.5b00489 BindingDB Entry DOI: 10.7270/Q2MP551H | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli) | BDBM50423645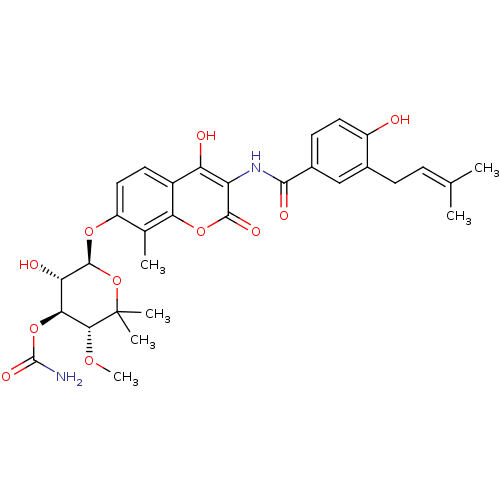 (Albamycin | Cathomycin | NOVOBIOCIN) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | n/a | 28 | n/a | n/a | n/a | n/a | n/a |
University of Ljubljana Curated by ChEMBL | Assay Description Binding affinity to Escherichia coli GyrB | J Med Chem 54: 915-29 (2011) Article DOI: 10.1021/jm101121s BindingDB Entry DOI: 10.7270/Q2NS0W23 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit A/B (Escherichia coli (strain K12)) | BDBM50095148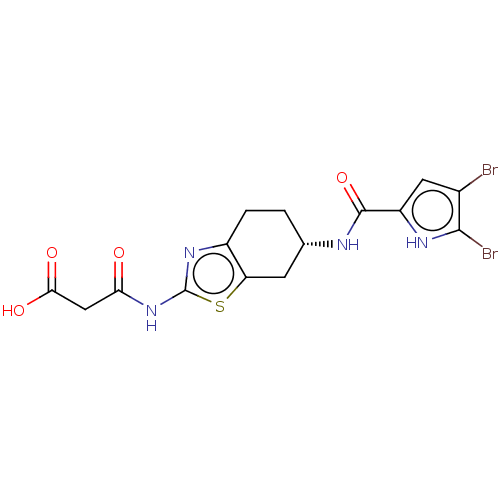 (CHEMBL3589535) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL KEGG PC cid PC sid UniChem Patents Similars | Article PubMed | n/a | n/a | n/a | 42 | n/a | n/a | n/a | n/a | n/a |
University of Ljubljana Curated by ChEMBL | Assay Description Binding affinity to Escherichia coli DNA gyrase subunit B N-terminal 24 kDa domain by SPR assay | J Med Chem 58: 5501-21 (2015) Article DOI: 10.1021/acs.jmedchem.5b00489 BindingDB Entry DOI: 10.7270/Q2MP551H | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit A/B (Escherichia coli (strain K12)) | BDBM50095211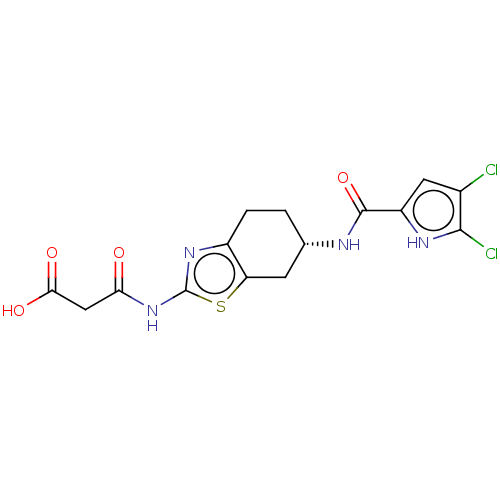 (CHEMBL3589079) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL KEGG PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | n/a | 49 | n/a | n/a | n/a | n/a | n/a |
University of Ljubljana Curated by ChEMBL | Assay Description Binding affinity to Escherichia coli DNA gyrase subunit B N-terminal 24 kDa domain by SPR assay | J Med Chem 58: 5501-21 (2015) Article DOI: 10.1021/acs.jmedchem.5b00489 BindingDB Entry DOI: 10.7270/Q2MP551H | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit A/B (Escherichia coli (strain K12)) | BDBM50095208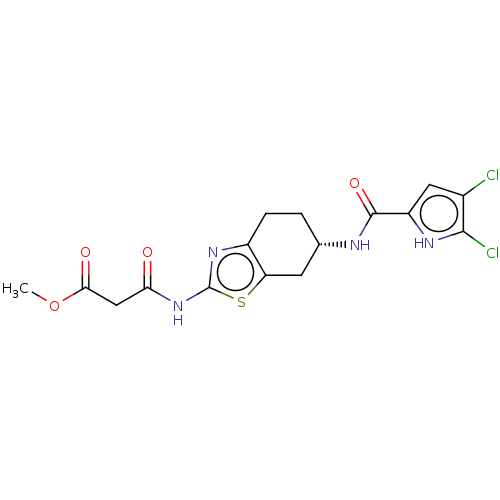 (CHEMBL3589076) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | n/a | 50 | n/a | n/a | n/a | n/a | n/a |
University of Ljubljana Curated by ChEMBL | Assay Description Binding affinity to Escherichia coli DNA gyrase subunit B N-terminal 24 kDa domain by SPR assay | J Med Chem 58: 5501-21 (2015) Article DOI: 10.1021/acs.jmedchem.5b00489 BindingDB Entry DOI: 10.7270/Q2MP551H | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit A/B (Escherichia coli (strain K12)) | BDBM50095254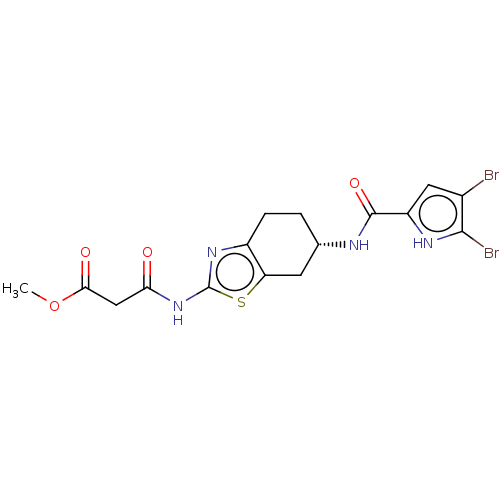 (CHEMBL3589530) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | n/a | 72 | n/a | n/a | n/a | n/a | n/a |
University of Ljubljana Curated by ChEMBL | Assay Description Binding affinity to Escherichia coli DNA gyrase subunit B N-terminal 24 kDa domain by SPR assay | J Med Chem 58: 5501-21 (2015) Article DOI: 10.1021/acs.jmedchem.5b00489 BindingDB Entry DOI: 10.7270/Q2MP551H | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit A/B (Escherichia coli (strain K12)) | BDBM50095290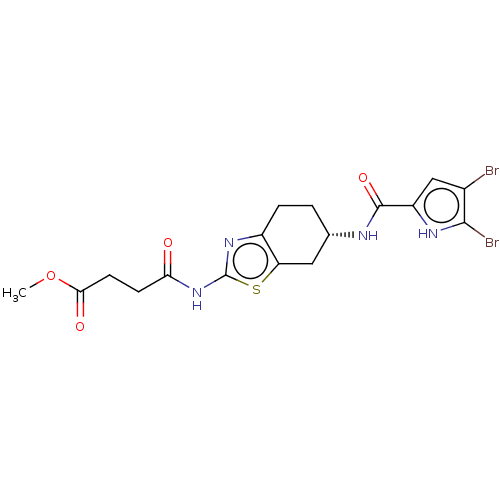 (CHEMBL3589531) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents | Article PubMed | n/a | n/a | n/a | 150 | n/a | n/a | n/a | n/a | n/a |
University of Ljubljana Curated by ChEMBL | Assay Description Binding affinity to Escherichia coli DNA gyrase subunit B N-terminal 24 kDa domain by SPR assay | J Med Chem 58: 5501-21 (2015) Article DOI: 10.1021/acs.jmedchem.5b00489 BindingDB Entry DOI: 10.7270/Q2MP551H | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50198240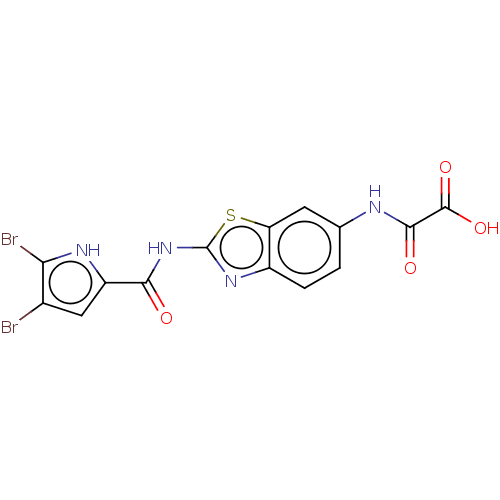 (CHEMBL3908487) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid PDB UniChem Similars | PDB Article PubMed | n/a | n/a | n/a | 230 | n/a | n/a | n/a | n/a | n/a |
University of Ljubljana Curated by ChEMBL | Assay Description Binding affinity to Escherichia coli DNA gyrase B ATP binding site by surface plasmon resonance assay | J Med Chem 59: 8941-8954 (2016) Article DOI: 10.1021/acs.jmedchem.6b00864 BindingDB Entry DOI: 10.7270/Q25B04GJ | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| DNA gyrase subunit A/B (Escherichia coli (strain K12)) | BDBM50095287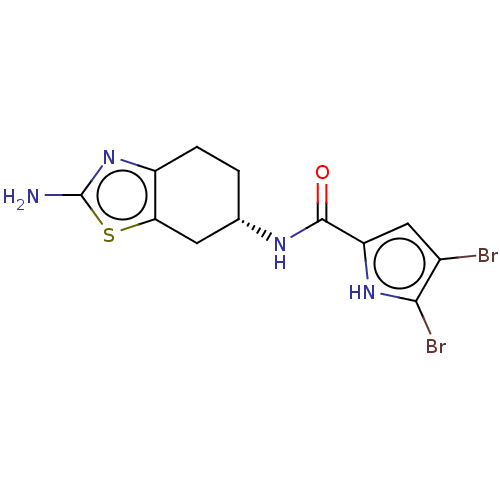 (CHEMBL3085880) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | n/a | 3.00E+3 | n/a | n/a | n/a | n/a | n/a |
University of Ljubljana Curated by ChEMBL | Assay Description Binding affinity to Escherichia coli DNA gyrase subunit B N-terminal 24 kDa domain by SPR assay | J Med Chem 58: 5501-21 (2015) Article DOI: 10.1021/acs.jmedchem.5b00489 BindingDB Entry DOI: 10.7270/Q2MP551H | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50259168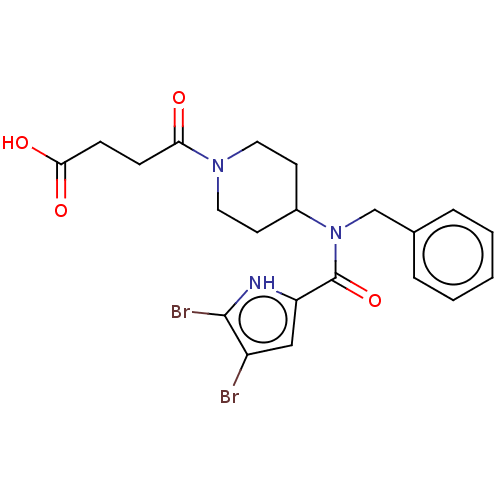 (CHEMBL4062117) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | n/a | 1.12E+4 | n/a | n/a | n/a | n/a | n/a |
University of Ljubljana Curated by ChEMBL | Assay Description Binding affinity to Escherichia coli ATCC 25922 DNA gyrase B by surface plasmon resonance assay | Eur J Med Chem 125: 500-514 (2017) Article DOI: 10.1016/j.ejmech.2016.09.040 BindingDB Entry DOI: 10.7270/Q27W6FNX | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50259169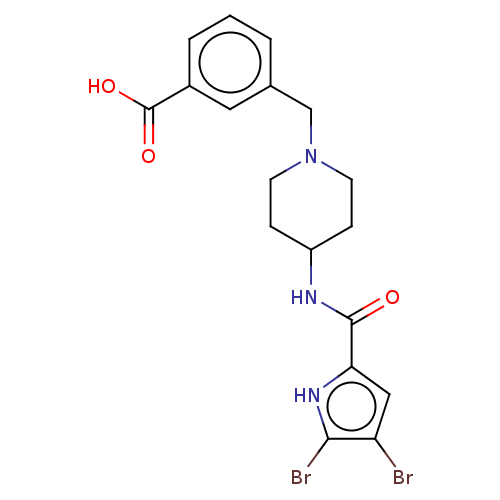 (CHEMBL4075133) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | n/a | 1.12E+4 | n/a | n/a | n/a | n/a | n/a |
University of Ljubljana Curated by ChEMBL | Assay Description Binding affinity to Escherichia coli ATCC 25922 DNA gyrase B by surface plasmon resonance assay | Eur J Med Chem 125: 500-514 (2017) Article DOI: 10.1016/j.ejmech.2016.09.040 BindingDB Entry DOI: 10.7270/Q27W6FNX | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50259163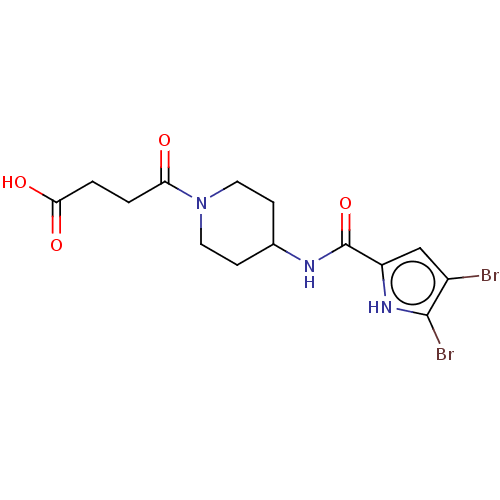 (CHEMBL4083521) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | n/a | 1.26E+4 | n/a | n/a | n/a | n/a | n/a |
University of Ljubljana Curated by ChEMBL | Assay Description Binding affinity to Escherichia coli ATCC 25922 DNA gyrase B by surface plasmon resonance assay | Eur J Med Chem 125: 500-514 (2017) Article DOI: 10.1016/j.ejmech.2016.09.040 BindingDB Entry DOI: 10.7270/Q27W6FNX | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit A/B (Escherichia coli (strain K12)) | BDBM50594763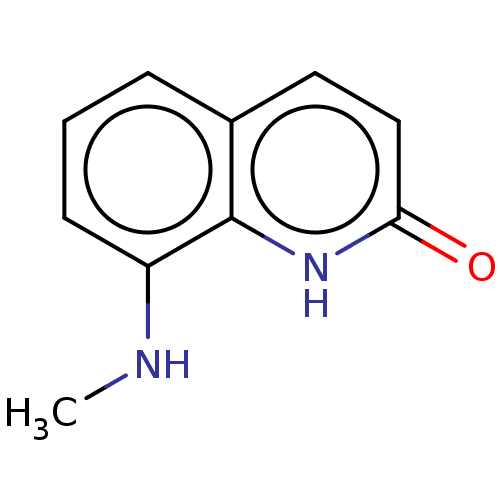 (CHEMBL5182050) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | n/a | 3.20E+4 | n/a | n/a | n/a | n/a | n/a |
TBA | Citation and Details Article DOI: 10.1021/acs.jmedchem.1c01803 BindingDB Entry DOI: 10.7270/Q2KD22XQ | ||||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50259170 (CHEMBL4103680) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | n/a | 2.56E+5 | n/a | n/a | n/a | n/a | n/a |
University of Ljubljana Curated by ChEMBL | Assay Description Binding affinity to Escherichia coli ATCC 25922 DNA gyrase B by surface plasmon resonance assay | Eur J Med Chem 125: 500-514 (2017) Article DOI: 10.1016/j.ejmech.2016.09.040 BindingDB Entry DOI: 10.7270/Q27W6FNX | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50408204 (CHEMBL4165664) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid PDB UniChem Similars | PDB Article PubMed | <0.0500 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
University of Ljubljana Curated by ChEMBL | Assay Description Inhibition of Escherichia coli DNA gyrase B | Eur J Med Chem 154: 117-132 (2018) Article DOI: 10.1016/j.ejmech.2018.05.011 BindingDB Entry DOI: 10.7270/Q2CV4M9N | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| DNA gyrase subunit A/B (Escherichia coli (strain K12)) | BDBM24611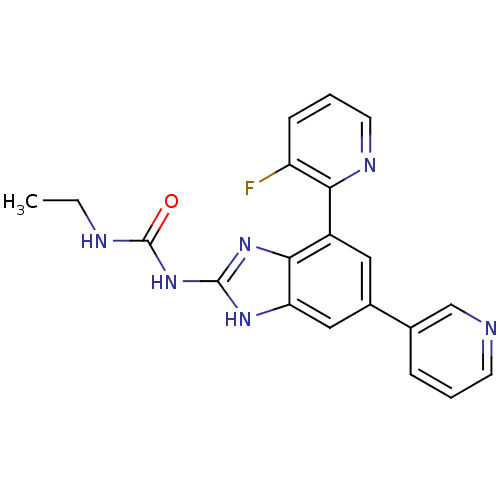 (3-ethyl-1-[7-(3-fluoropyridin-2-yl)-5-(pyridin-3-y...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | <4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Vertex | Assay Description Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ... | J Med Chem 51: 5243-63 (2008) Article DOI: 10.1021/jm800318d BindingDB Entry DOI: 10.7270/Q2J67F7T | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit A/B (Escherichia coli (strain K12)) | BDBM24606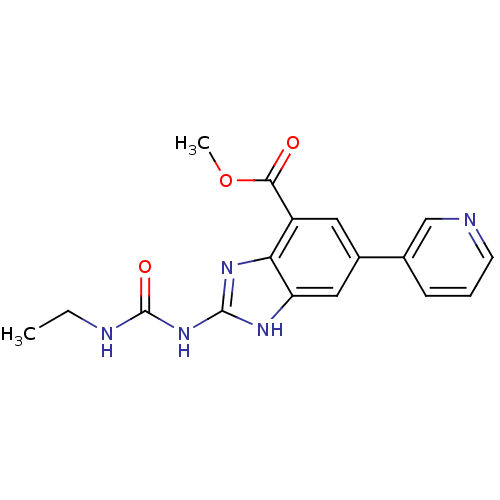 (Benzimidazole urea analogue, 10 | methyl 2-[(ethyl...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Patents | Article PubMed | <4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Vertex | Assay Description Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ... | J Med Chem 51: 5243-63 (2008) Article DOI: 10.1021/jm800318d BindingDB Entry DOI: 10.7270/Q2J67F7T | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit A/B (Escherichia coli (strain K12)) | BDBM24608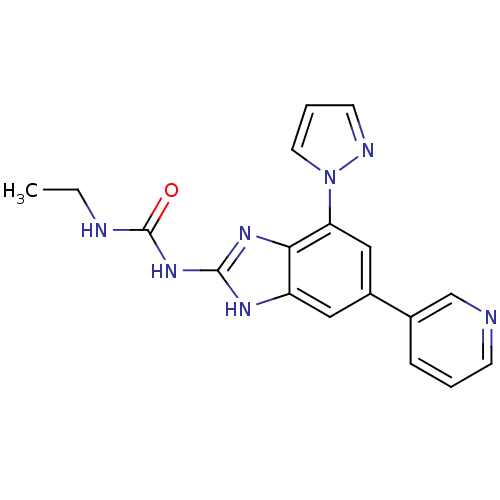 (3-ethyl-1-[7-(1H-pyrazol-1-yl)-5-(pyridin-3-yl)-1H...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | <4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Vertex | Assay Description Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ... | J Med Chem 51: 5243-63 (2008) Article DOI: 10.1021/jm800318d BindingDB Entry DOI: 10.7270/Q2J67F7T | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit A/B (Escherichia coli (strain K12)) | BDBM24609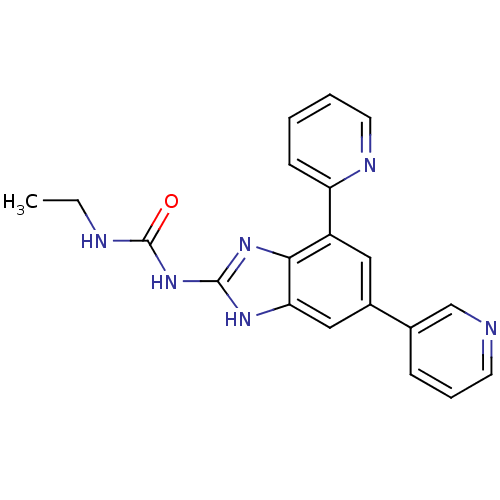 (3-ethyl-1-[7-(pyridin-2-yl)-5-(pyridin-3-yl)-1H-1,...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | <4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Vertex | Assay Description Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ... | J Med Chem 51: 5243-63 (2008) Article DOI: 10.1021/jm800318d BindingDB Entry DOI: 10.7270/Q2J67F7T | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit A/B (Escherichia coli (strain K12)) | BDBM24601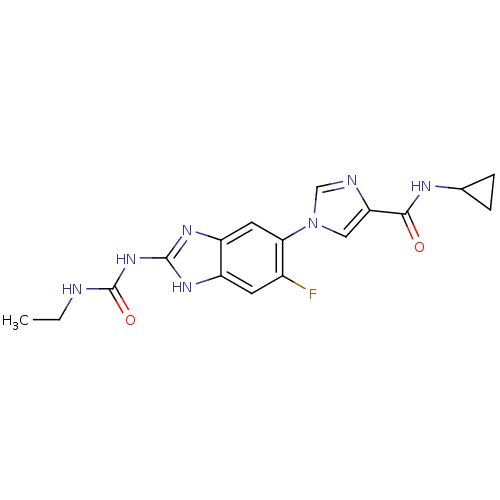 (Benzimidazole urea analogue, 5 | N-cyclopropyl-1-{...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Patents | Article PubMed | <4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Vertex | Assay Description Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ... | J Med Chem 51: 5243-63 (2008) Article DOI: 10.1021/jm800318d BindingDB Entry DOI: 10.7270/Q2J67F7T | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit A/B (Escherichia coli (strain K12)) | BDBM24617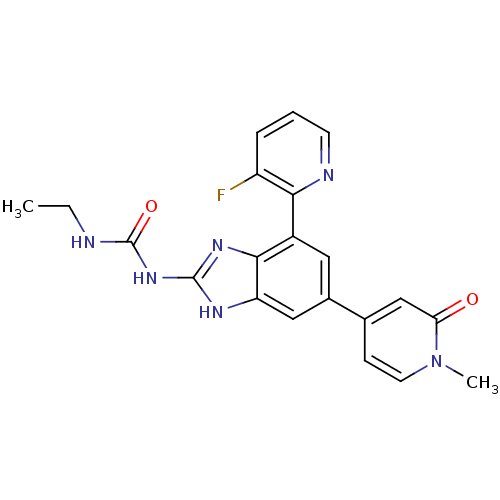 (3-ethyl-1-[7-(3-fluoropyridin-2-yl)-5-(1-methyl-2-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | <4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Vertex | Assay Description Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ... | J Med Chem 51: 5243-63 (2008) Article DOI: 10.1021/jm800318d BindingDB Entry DOI: 10.7270/Q2J67F7T | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit A/B (Escherichia coli (strain K12)) | BDBM24614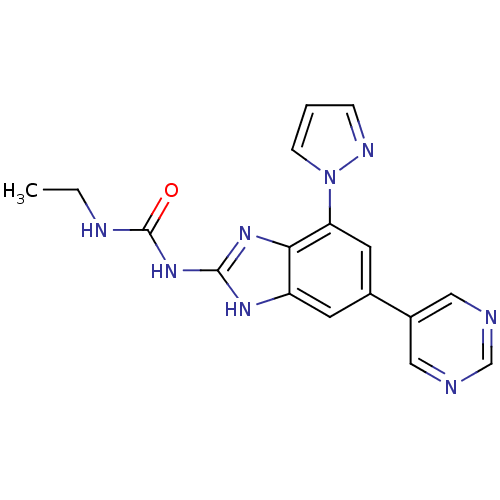 (3-ethyl-1-[7-(1H-pyrazol-1-yl)-5-(pyrimidin-5-yl)-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | <4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Vertex | Assay Description Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ... | J Med Chem 51: 5243-63 (2008) Article DOI: 10.1021/jm800318d BindingDB Entry DOI: 10.7270/Q2J67F7T | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit A/B (Escherichia coli (strain K12)) | BDBM24615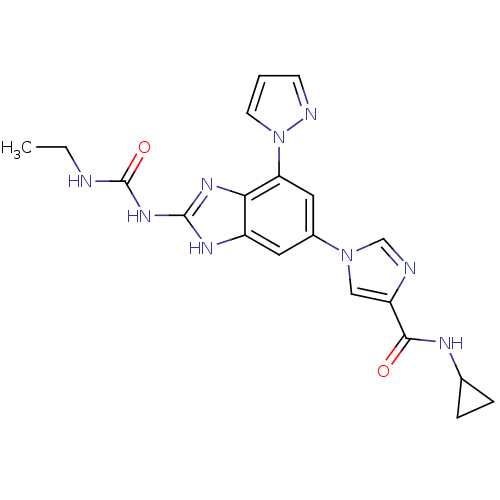 (Benzimidazole urea analogue, 19 | N-cyclopropyl-1-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Patents | Article PubMed | <4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Vertex | Assay Description Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ... | J Med Chem 51: 5243-63 (2008) Article DOI: 10.1021/jm800318d BindingDB Entry DOI: 10.7270/Q2J67F7T | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit A/B (Escherichia coli (strain K12)) | BDBM24616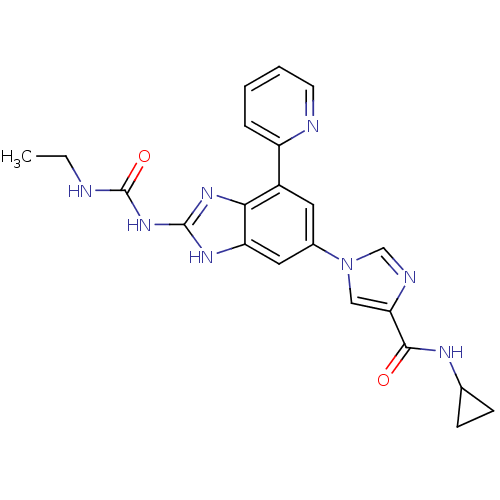 (1-(6-(4-(Cyclopropylcarbamoyl)-1H-imidazol-1-yl)-4...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Patents | Article PubMed | <4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Vertex | Assay Description Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ... | J Med Chem 51: 5243-63 (2008) Article DOI: 10.1021/jm800318d BindingDB Entry DOI: 10.7270/Q2J67F7T | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit A/B (Escherichia coli (strain K12)) | BDBM50497604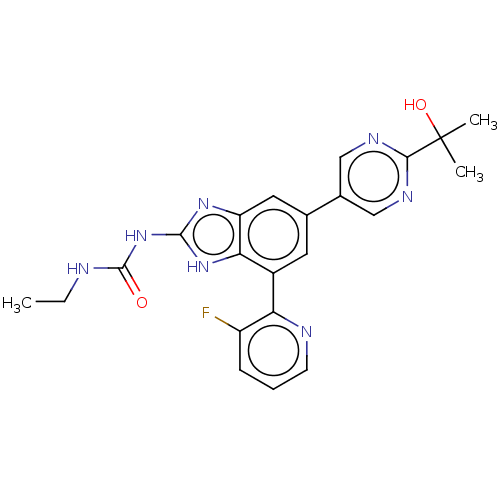 (CHEMBL3264033) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | <4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Vertex Pharmaceuticals Inc Curated by ChEMBL | Assay Description Inhibition of aureus Escherichia coli DNA gyrase A2B2 using pBR322 plasmid DNA as substrate by coupled enzyme reaction assay | J Med Chem 57: 8792-816 (2014) Article DOI: 10.1021/jm500563g BindingDB Entry DOI: 10.7270/Q2TF01BF | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit A/B (Escherichia coli (strain K12)) | BDBM50497603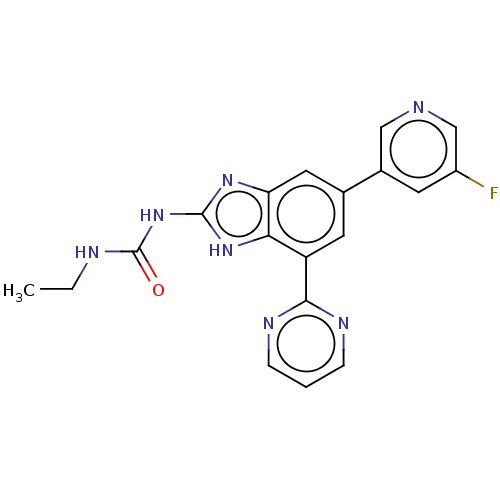 (CHEMBL3356986) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid PDB UniChem | Article PubMed | 5 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Vertex Pharmaceuticals Inc Curated by ChEMBL | Assay Description Inhibition of aureus Escherichia coli DNA gyrase A2B2 using pBR322 plasmid DNA as substrate by coupled enzyme reaction assay | J Med Chem 57: 8792-816 (2014) Article DOI: 10.1021/jm500563g BindingDB Entry DOI: 10.7270/Q2TF01BF | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit A/B (Escherichia coli (strain K12)) | BDBM24607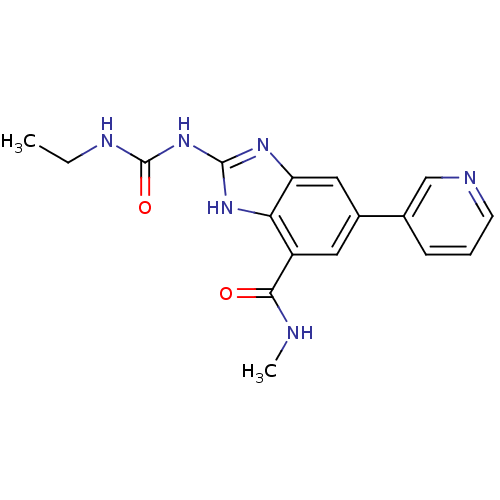 (2-[(ethylcarbamoyl)amino]-N-methyl-5-(pyridin-3-yl...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Patents | Article PubMed | 5 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Vertex | Assay Description Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ... | J Med Chem 51: 5243-63 (2008) Article DOI: 10.1021/jm800318d BindingDB Entry DOI: 10.7270/Q2J67F7T | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit A/B (Escherichia coli (strain K12)) | BDBM50497602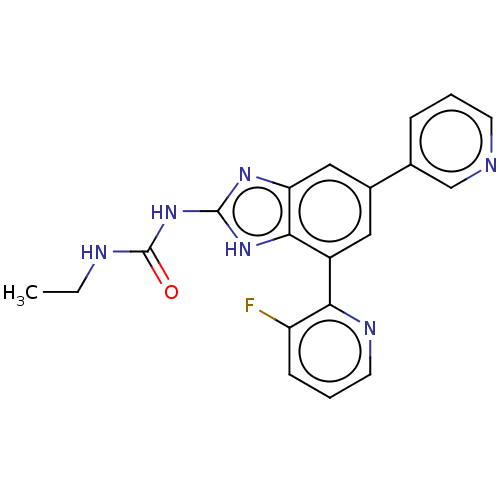 (CHEMBL222333 | VRT-752586) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 6 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Vertex Pharmaceuticals Inc Curated by ChEMBL | Assay Description Inhibition of aureus Escherichia coli DNA gyrase A2B2 using pBR322 plasmid DNA as substrate by coupled enzyme reaction assay | J Med Chem 57: 8792-816 (2014) Article DOI: 10.1021/jm500563g BindingDB Entry DOI: 10.7270/Q2TF01BF | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit A/B (Escherichia coli (strain K12)) | BDBM24610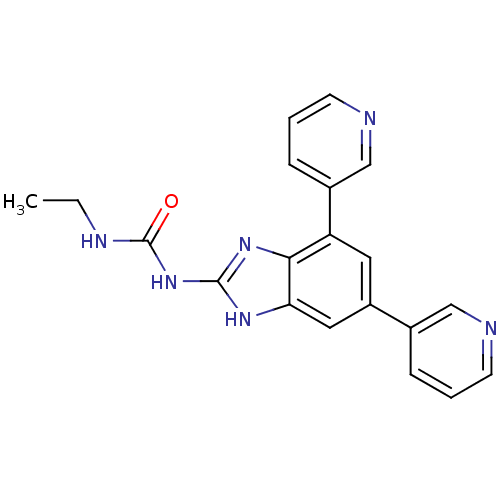 (1-[5,7-bis(pyridin-3-yl)-1H-1,3-benzodiazol-2-yl]-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 6 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Vertex | Assay Description Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ... | J Med Chem 51: 5243-63 (2008) Article DOI: 10.1021/jm800318d BindingDB Entry DOI: 10.7270/Q2J67F7T | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50226181 ((3R,4S,5R,6R)-5-hydroxy-6-(4-hydroxy-3-(4-hydroxy-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE KEGG MMDB PC cid PC sid PDB UniChem Similars | MMDB Article PubMed | 7 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
R. C. Patel Institute of Pharmaceutical Education and Research Curated by ChEMBL | Assay Description Inhibition of Escherichia coli H560 DNA gyrase B ATPase activity after 30 mins by ammonium molybdate/malachite green-based phosphate detection assay | Eur J Med Chem 124: 160-185 (2016) Article DOI: 10.1016/j.ejmech.2016.08.034 BindingDB Entry DOI: 10.7270/Q2K64M38 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit A/B (Staphylococcus aureus) | BDBM24609 (3-ethyl-1-[7-(pyridin-2-yl)-5-(pyridin-3-yl)-1H-1,...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 7 | -47.3 | n/a | n/a | n/a | n/a | n/a | 7.5 | 30 |
Vertex | Assay Description Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ... | J Med Chem 51: 5243-63 (2008) Article DOI: 10.1021/jm800318d BindingDB Entry DOI: 10.7270/Q2J67F7T | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit A/B (Staphylococcus aureus) | BDBM24606 (Benzimidazole urea analogue, 10 | methyl 2-[(ethyl...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Patents | Article PubMed | 8 | -47.0 | n/a | n/a | n/a | n/a | n/a | 7.5 | 30 |
Vertex | Assay Description Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ... | J Med Chem 51: 5243-63 (2008) Article DOI: 10.1021/jm800318d BindingDB Entry DOI: 10.7270/Q2J67F7T | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Staphylococcus aureus) | BDBM50393079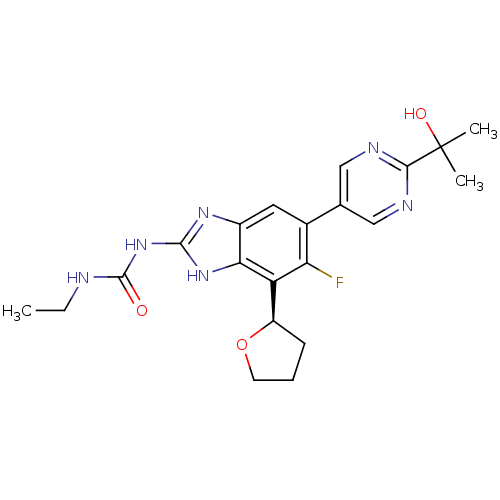 (CHEMBL2152855 | US9040542, 23) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid UniChem Similars | Article PubMed | 9 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA Curated by ChEMBL | Assay Description Inhibition of Staphylococcus aureus DNA gyrase Gyr B subunit by ATPase assay | ACS Med Chem Lett 3: 783-784 (2012) Article DOI: 10.1021/ml300234y BindingDB Entry DOI: 10.7270/Q2N29Z1N | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit A/B (Staphylococcus aureus) | BDBM50393079 (CHEMBL2152855 | US9040542, 23) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid UniChem Similars | US Patent | 9 | n/a | n/a | n/a | n/a | n/a | n/a | 7.6 | n/a |
VERTEX PHARMACEUTICALS INCORPORATED US Patent | Assay Description The ATP hydrolysis activity of S. aureus DNA gyrase is measured by coupling the production of ADP through pyruvate kinase/lactate dehydrogenase to th... | US Patent US9040542 (2015) BindingDB Entry DOI: 10.7270/Q2F18XHX | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit A/B (Staphylococcus aureus) | BDBM50393079 (CHEMBL2152855 | US9040542, 23) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid UniChem Similars | Article PubMed | 9 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Vertex Pharmaceuticals Inc Curated by ChEMBL | Assay Description Inhibition of Staphylococcus aureus DNA gyrase using pBR322 plasmid DNA as substrate by coupled enzyme reaction assay | J Med Chem 57: 8792-816 (2014) Article DOI: 10.1021/jm500563g BindingDB Entry DOI: 10.7270/Q2TF01BF | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit A/B (Staphylococcus aureus) | BDBM24601 (Benzimidazole urea analogue, 5 | N-cyclopropyl-1-{...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Patents | Article PubMed | 10 | -46.4 | n/a | n/a | n/a | n/a | n/a | 7.5 | 30 |
Vertex | Assay Description Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ... | J Med Chem 51: 5243-63 (2008) Article DOI: 10.1021/jm800318d BindingDB Entry DOI: 10.7270/Q2J67F7T | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit A/B (Staphylococcus aureus) | BDBM282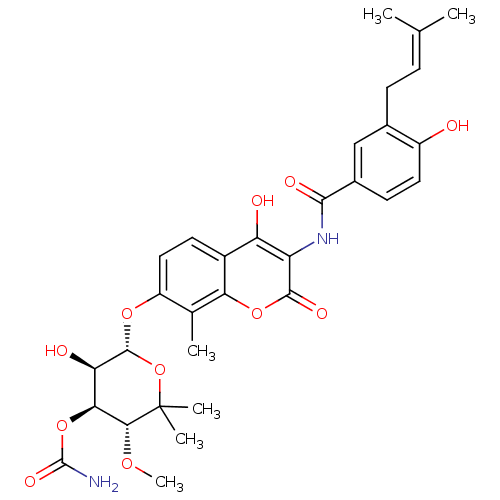 ((3R,4S,5R,6R)-5-hydroxy-6-[(2-hydroxy-3-{[4-hydrox...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase DrugBank MCE KEGG MMDB PC cid PC sid PDB UniChem Similars | DrugBank PDB Article PubMed | 10 | -46.4 | n/a | n/a | n/a | n/a | n/a | 7.5 | 30 |
Vertex | Assay Description Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ... | J Med Chem 51: 5243-63 (2008) Article DOI: 10.1021/jm800318d BindingDB Entry DOI: 10.7270/Q2J67F7T | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| DNA gyrase subunit A/B (Staphylococcus aureus) | BDBM24615 (Benzimidazole urea analogue, 19 | N-cyclopropyl-1-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Patents | Article PubMed | 10 | -46.4 | n/a | n/a | n/a | n/a | n/a | 7.5 | 30 |
Vertex | Assay Description Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ... | J Med Chem 51: 5243-63 (2008) Article DOI: 10.1021/jm800318d BindingDB Entry DOI: 10.7270/Q2J67F7T | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit A/B (Staphylococcus aureus) | BDBM24616 (1-(6-(4-(Cyclopropylcarbamoyl)-1H-imidazol-1-yl)-4...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Patents | Article PubMed | 12 | -46.0 | n/a | n/a | n/a | n/a | n/a | 7.5 | 30 |
Vertex | Assay Description Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ... | J Med Chem 51: 5243-63 (2008) Article DOI: 10.1021/jm800318d BindingDB Entry DOI: 10.7270/Q2J67F7T | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit A/B (Staphylococcus aureus) | BDBM24611 (3-ethyl-1-[7-(3-fluoropyridin-2-yl)-5-(pyridin-3-y...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 14 | -45.6 | n/a | n/a | n/a | n/a | n/a | 7.5 | 30 |
Vertex | Assay Description Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ... | J Med Chem 51: 5243-63 (2008) Article DOI: 10.1021/jm800318d BindingDB Entry DOI: 10.7270/Q2J67F7T | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit A/B (Escherichia coli (strain K12)) | BDBM282 ((3R,4S,5R,6R)-5-hydroxy-6-[(2-hydroxy-3-{[4-hydrox...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase DrugBank MCE KEGG MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | 14 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Vertex | Assay Description Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ... | J Med Chem 51: 5243-63 (2008) Article DOI: 10.1021/jm800318d BindingDB Entry DOI: 10.7270/Q2J67F7T | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit A/B (Escherichia coli (strain K12)) | BDBM24613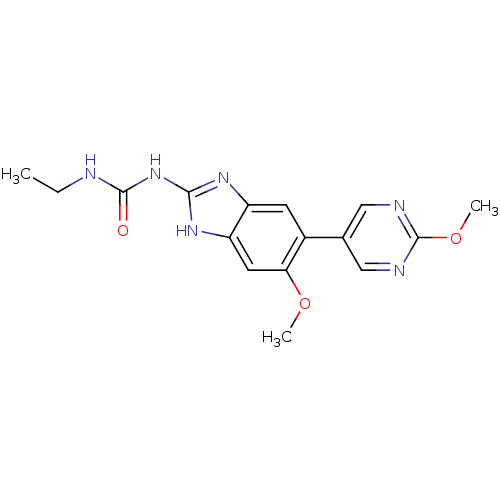 (3-ethyl-1-[6-methoxy-5-(2-methoxypyrimidin-5-yl)-1...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 15 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Vertex | Assay Description Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ... | J Med Chem 51: 5243-63 (2008) Article DOI: 10.1021/jm800318d BindingDB Entry DOI: 10.7270/Q2J67F7T | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit A/B (Staphylococcus aureus) | BDBM24608 (3-ethyl-1-[7-(1H-pyrazol-1-yl)-5-(pyridin-3-yl)-1H...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 15 | -45.4 | n/a | n/a | n/a | n/a | n/a | 7.5 | 30 |
Vertex | Assay Description Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ... | J Med Chem 51: 5243-63 (2008) Article DOI: 10.1021/jm800318d BindingDB Entry DOI: 10.7270/Q2J67F7T | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit A/B (Staphylococcus aureus) | BDBM24614 (3-ethyl-1-[7-(1H-pyrazol-1-yl)-5-(pyrimidin-5-yl)-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 16 | -45.2 | n/a | n/a | n/a | n/a | n/a | 7.5 | 30 |
Vertex | Assay Description Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ... | J Med Chem 51: 5243-63 (2008) Article DOI: 10.1021/jm800318d BindingDB Entry DOI: 10.7270/Q2J67F7T | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit A/B (Staphylococcus aureus) | BDBM24610 (1-[5,7-bis(pyridin-3-yl)-1H-1,3-benzodiazol-2-yl]-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 17 | -45.1 | n/a | n/a | n/a | n/a | n/a | 7.5 | 30 |
Vertex | Assay Description Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ... | J Med Chem 51: 5243-63 (2008) Article DOI: 10.1021/jm800318d BindingDB Entry DOI: 10.7270/Q2J67F7T | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit A/B (Staphylococcus aureus) | BDBM50112815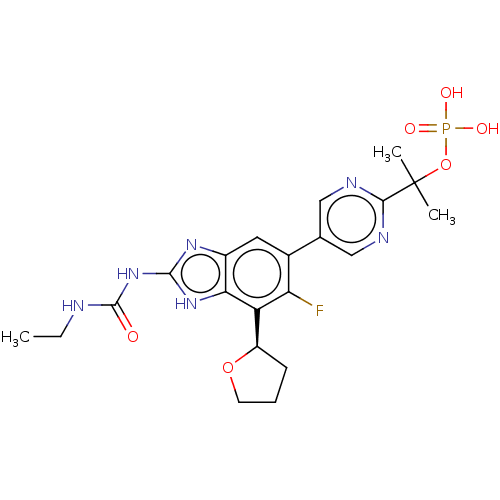 (CHEMBL2221212) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid UniChem Similars | Article PubMed | <17.7 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Vertex Pharmaceuticals Inc Curated by ChEMBL | Assay Description Inhibition of Staphylococcus aureus DNA gyrase | ACS Med Chem Lett 6: 822-6 (2015) Article DOI: 10.1021/acsmedchemlett.5b00196 BindingDB Entry DOI: 10.7270/Q2J67JPW | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit A/B (Escherichia coli (strain K12)) | BDBM24612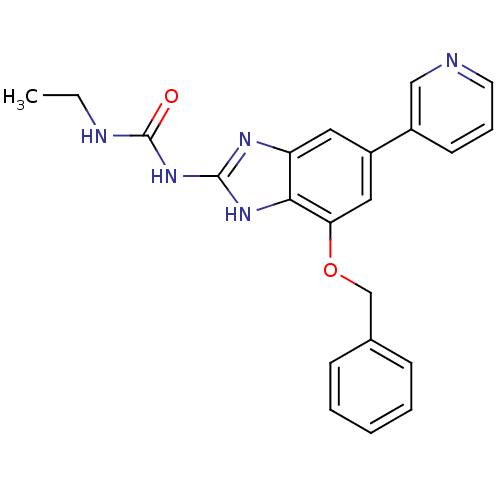 (1-[7-(benzyloxy)-5-(pyridin-3-yl)-1H-1,3-benzodiaz...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Patents | Article PubMed | 18 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Vertex | Assay Description Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ... | J Med Chem 51: 5243-63 (2008) Article DOI: 10.1021/jm800318d BindingDB Entry DOI: 10.7270/Q2J67F7T | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit A/B (Escherichia coli (strain K12)) | BDBM24600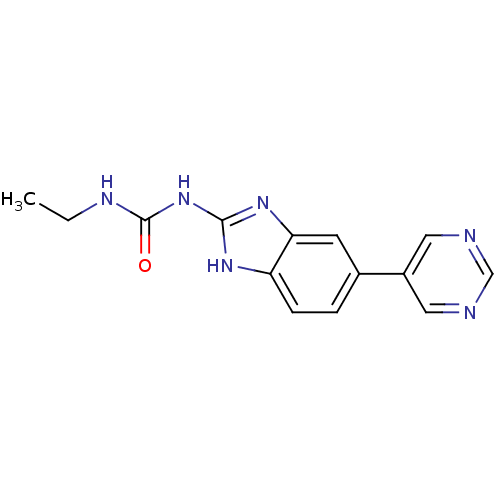 (3-ethyl-1-[5-(pyrimidin-5-yl)-1H-1,3-benzodiazol-2...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 20 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Vertex | Assay Description Enzymatic hydrolysis of ATP to ADP was coupled to the conversion of NADH to NAD+. The decrease in NADH absorbance was monitored at 340 nm for 20 min ... | J Med Chem 51: 5243-63 (2008) Article DOI: 10.1021/jm800318d BindingDB Entry DOI: 10.7270/Q2J67F7T | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Displayed 1 to 50 (of 2713 total ) | Next | Last >> |