

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Reaction Details | |||
|---|---|---|---|
 | Report a problem with these data | ||
| Target | Cytochrome P450 3A4 | ||
| Ligand | BDBM50153594 | ||
| Substrate/Competitor | n/a | ||
| Meas. Tech. | ChEMBL_1561100 (CHEMBL3776824) | ||
| IC50 | >50000±n/a nM | ||
| Citation |  Hirata, K; Kotoku, M; Seki, N; Maeba, T; Maeda, K; Hirashima, S; Sakai, T; Obika, S; Hori, A; Hase, Y; Yamaguchi, T; Katsuda, Y; Hata, T; Miyagawa, N; Arita, K; Nomura, Y; Asahina, K; Aratsu, Y; Kamada, M; Adachi, T; Noguchi, M; Doi, S; Crowe, P; Bradley, E; Steensma, R; Tao, H; Fenn, M; Babine, R; Li, X; Thacher, S; Hashimoto, H; Shiozaki, M SAR Exploration Guided by LE and Fsp(3): Discovery of a Selective and Orally Efficacious ROR� Inhibitor. ACS Med Chem Lett7:23-7 (2016) [PubMed] Article Hirata, K; Kotoku, M; Seki, N; Maeba, T; Maeda, K; Hirashima, S; Sakai, T; Obika, S; Hori, A; Hase, Y; Yamaguchi, T; Katsuda, Y; Hata, T; Miyagawa, N; Arita, K; Nomura, Y; Asahina, K; Aratsu, Y; Kamada, M; Adachi, T; Noguchi, M; Doi, S; Crowe, P; Bradley, E; Steensma, R; Tao, H; Fenn, M; Babine, R; Li, X; Thacher, S; Hashimoto, H; Shiozaki, M SAR Exploration Guided by LE and Fsp(3): Discovery of a Selective and Orally Efficacious ROR� Inhibitor. ACS Med Chem Lett7:23-7 (2016) [PubMed] Article | ||
| More Info.: | Get all data from this article, Assay Method | ||
| Cytochrome P450 3A4 | |||
| Name: | Cytochrome P450 3A4 | ||
| Synonyms: | Albendazole monooxygenase | Albendazole sulfoxidase | CP3A4_HUMAN | CYP3A3 | CYP3A4 | CYPIIIA3 | CYPIIIA4 | Cytochrome P450 3A3 | Cytochrome P450 3A4 (CYP3A4) | Cytochrome P450 HLp | Nifedipine oxidase | Quinine 3-monooxygenase | Taurochenodeoxycholate 6-alpha-hydroxylase | ||
| Type: | Enzyme | ||
| Mol. Mass.: | 57349.57 | ||
| Organism: | Homo sapiens (Human) | ||
| Description: | n/a | ||
| Residue: | 503 | ||
| Sequence: |
| ||
| BDBM50153594 | |||
| n/a | |||
| Name | BDBM50153594 | ||
| Synonyms: | CHEMBL3774855 | ||
| Type | Small organic molecule | ||
| Emp. Form. | C28H39N5O2 | ||
| Mol. Mass. | 477.6416 | ||
| SMILES | CC(C)C[C@H]1C[C@H](C1)c1nnc([C@H]2CN(C[C@@H]2C(=O)Nc2ccc(C)cc2C)C(C)=O)n1C1CC1 |r,wU:4.3,6.8,12.12,wD:16.18,(2.36,7.33,;2.68,6.14,;1.81,5.27,;4.17,5.74,;4.56,4.26,;5.9,3.48,;5.11,2.21,;3.79,2.92,;5.62,.75,;7.1,.32,;7.14,-1.22,;5.7,-1.71,;5.38,-3.22,;6.42,-4.36,;5.65,-5.7,;4.14,-5.38,;4,-3.86,;2.66,-3.08,;2.67,-1.85,;1.33,-3.85,;-.01,-3.08,;0,-1.54,;-1.33,-.76,;-2.67,-1.53,;-3.73,-.91,;-2.67,-3.07,;-1.34,-3.84,;-1.35,-5.07,;6.27,-7.1,;5.55,-8.1,;7.5,-7.23,;4.75,-.52,;3.21,-.57,;1.93,-1.26,;2.03,.28,)| | ||
| Structure | 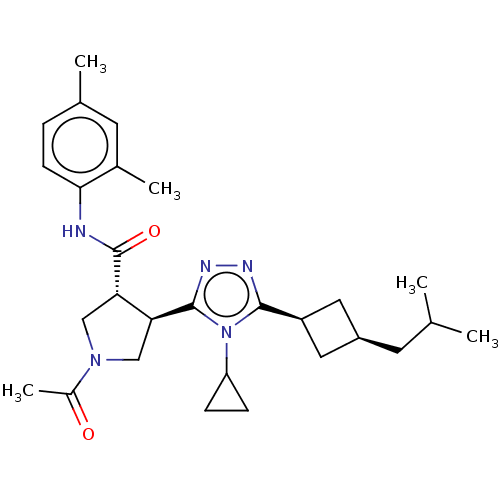
| ||