| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Oxysterols receptor LXR-alpha |
|---|
| Ligand | BDBM50541922 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_1985560 (CHEMBL4618966) |
|---|
| EC50 | >7500±n/a nM |
|---|
| Citation |  Cherney, RJ; Cornelius, LAM; Srivastava, A; Weigelt, CA; Marcoux, D; Duan, JJ; Shi, Q; Batt, DG; Liu, Q; Yip, S; Wu, DR; Ruzanov, M; Sack, J; Khan, J; Wang, J; Yarde, M; Cvijic, ME; Mathur, A; Li, S; Shuster, D; Khandelwal, P; Borowski, V; Xie, J; Obermeier, M; Fura, A; Stefanski, K; Cornelius, G; Tino, JA; Macor, JE; Salter-Cid, L; Denton, R; Zhao, Q; Carter, PH; Dhar, TGM Discovery of BMS-986251: A Clinically Viable, Potent, and Selective ROR?t Inverse Agonist. ACS Med Chem Lett11:1221-1227 (2020) [PubMed] Article Cherney, RJ; Cornelius, LAM; Srivastava, A; Weigelt, CA; Marcoux, D; Duan, JJ; Shi, Q; Batt, DG; Liu, Q; Yip, S; Wu, DR; Ruzanov, M; Sack, J; Khan, J; Wang, J; Yarde, M; Cvijic, ME; Mathur, A; Li, S; Shuster, D; Khandelwal, P; Borowski, V; Xie, J; Obermeier, M; Fura, A; Stefanski, K; Cornelius, G; Tino, JA; Macor, JE; Salter-Cid, L; Denton, R; Zhao, Q; Carter, PH; Dhar, TGM Discovery of BMS-986251: A Clinically Viable, Potent, and Selective ROR?t Inverse Agonist. ACS Med Chem Lett11:1221-1227 (2020) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Oxysterols receptor LXR-alpha |
|---|
| Name: | Oxysterols receptor LXR-alpha |
|---|
| Synonyms: | LXRA | Liver X Receptor alpha (LXR-alpha) | Liver X receptor alpha | Liver X receptor alpha (LXRA) | Liver X receptor alpha (NR1H3) | Liver X, LXR alpha | NR1H3 | NR1H3_HUMAN | Nuclear orphan receptor LXR-alpha | Nuclear receptor subfamily 1 group H member 3 |
|---|
| Type: | Enzyme Catalytic Domain |
|---|
| Mol. Mass.: | 50403.85 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q13133 |
|---|
| Residue: | 447 |
|---|
| Sequence: | MSLWLGAPVPDIPPDSAVELWKPGAQDASSQAQGGSSCILREEARMPHSAGGTAGVGLEA
AEPTALLTRAEPPSEPTEIRPQKRKKGPAPKMLGNELCSVCGDKASGFHYNVLSCEGCKG
FFRRSVIKGAHYICHSGGHCPMDTYMRRKCQECRLRKCRQAGMREECVLSEEQIRLKKLK
RQEEEQAHATSLPPRASSPPQILPQLSPEQLGMIEKLVAAQQQCNRRSFSDRLRVTPWPM
APDPHSREARQQRFAHFTELAIVSVQEIVDFAKQLPGFLQLSREDQIALLKTSAIEVMLL
ETSRRYNPGSESITFLKDFSYNREDFAKAGLQVEFINPIFEFSRAMNELQLNDAEFALLI
AISIFSADRPNVQDQLQVERLQHTYVEALHAYVSIHHPHDRLMFPRMLMKLVSLRTLSSV
HSEQVFALRLQDKKLPPLLSEIWDVHE
|
|
|
|---|
| BDBM50541922 |
|---|
| n/a |
|---|
| Name | BDBM50541922 |
|---|
| Synonyms: | Bms-986251 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C30H29F8NO5S |
|---|
| Mol. Mass. | 667.607 |
|---|
| SMILES | [H][C@@]12CCc3cc(ccc3[C@@]1(CCN2C(=O)[C@@H]1CC[C@H](C[C@@H]1C)C(O)=O)S(=O)(=O)c1ccc(F)cc1)C(F)(C(F)(F)F)C(F)(F)F |r| |
|---|
| Structure | 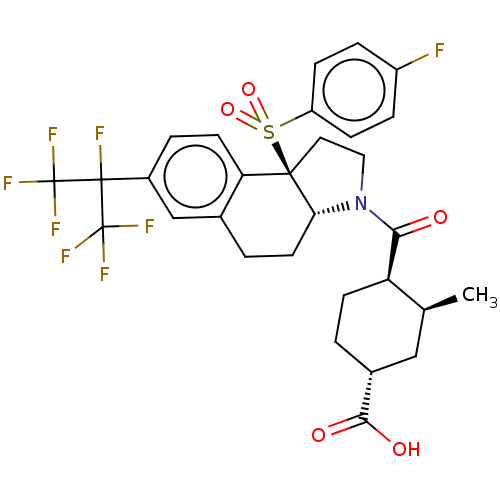
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Cherney, RJ; Cornelius, LAM; Srivastava, A; Weigelt, CA; Marcoux, D; Duan, JJ; Shi, Q; Batt, DG; Liu, Q; Yip, S; Wu, DR; Ruzanov, M; Sack, J; Khan, J; Wang, J; Yarde, M; Cvijic, ME; Mathur, A; Li, S; Shuster, D; Khandelwal, P; Borowski, V; Xie, J; Obermeier, M; Fura, A; Stefanski, K; Cornelius, G; Tino, JA; Macor, JE; Salter-Cid, L; Denton, R; Zhao, Q; Carter, PH; Dhar, TGM Discovery of BMS-986251: A Clinically Viable, Potent, and Selective ROR?t Inverse Agonist. ACS Med Chem Lett11:1221-1227 (2020) [PubMed] Article
Cherney, RJ; Cornelius, LAM; Srivastava, A; Weigelt, CA; Marcoux, D; Duan, JJ; Shi, Q; Batt, DG; Liu, Q; Yip, S; Wu, DR; Ruzanov, M; Sack, J; Khan, J; Wang, J; Yarde, M; Cvijic, ME; Mathur, A; Li, S; Shuster, D; Khandelwal, P; Borowski, V; Xie, J; Obermeier, M; Fura, A; Stefanski, K; Cornelius, G; Tino, JA; Macor, JE; Salter-Cid, L; Denton, R; Zhao, Q; Carter, PH; Dhar, TGM Discovery of BMS-986251: A Clinically Viable, Potent, and Selective ROR?t Inverse Agonist. ACS Med Chem Lett11:1221-1227 (2020) [PubMed] Article