

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trypsin (Homo sapiens (Human)) | BDBM50507483 (CHEMBL4530379) | PDB KEGG UniProtKB/SwissProt antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 0.00230 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The University of Queensland Curated by ChEMBL | Assay Description Inhibition of trypsin (unknown origin) | ACS Med Chem Lett 9: 1258-1262 (2018) Article DOI: 10.1021/acsmedchemlett.8b00422 BindingDB Entry DOI: 10.7270/Q2Q81HCD | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17122 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.00400 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17127 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.00400 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Gag-Pol polyprotein [489-587] (Human immunodeficiency virus type 1) | BDBM578 ((2S)-N-[(2S,4S,5S)-5-[2-(2,6-dimethylphenoxy)aceta...) | PDB MMDB UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL MCE MMDB PC cid PC sid PDB UniChem Patents Similars | MMDB PDB PubMed | 0.00400 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
University of Queensland Curated by ChEMBL | Assay Description Binding affinity against ritonavir-resistant strains. | J Med Chem 43: 305-41 (2000) Checked by Author BindingDB Entry DOI: 10.7270/Q2JD4XH4 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17129 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.00400 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17136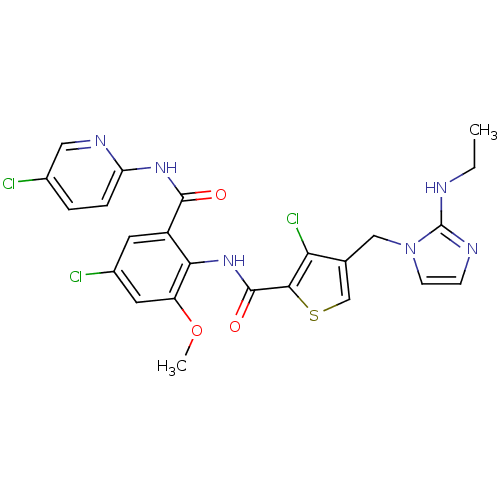 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.00500 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17135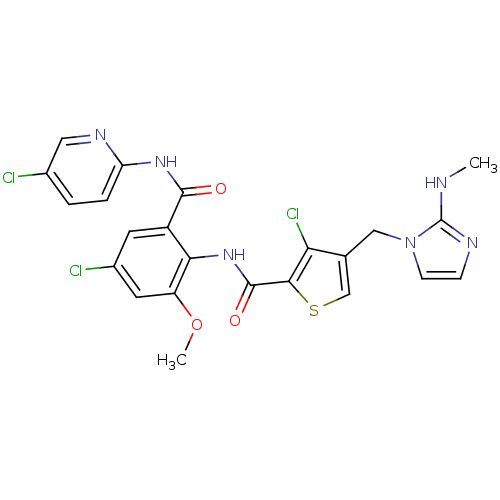 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | MMDB PC cid PC sid PDB UniChem Patents Similars | MMDB PDB Article PubMed | 0.00500 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Melanocyte-stimulating hormone receptor (Homo sapiens (Human)) | BDBM50590042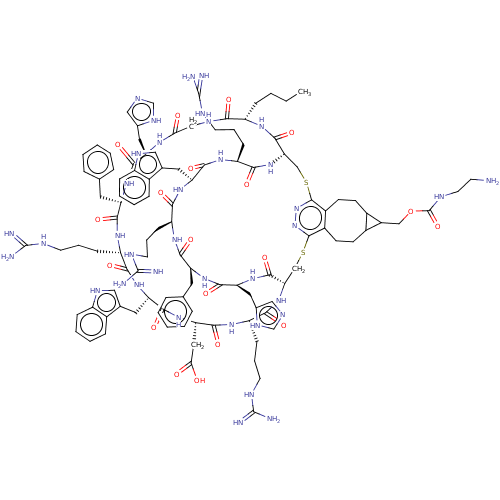 (CHEMBL5175981) | PDB KEGG UniProtKB/SwissProt antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | >0.00549 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA | Citation and Details Article DOI: 10.1021/acs.jmedchem.2c00793 BindingDB Entry DOI: 10.7270/Q20C50R5 | ||||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17134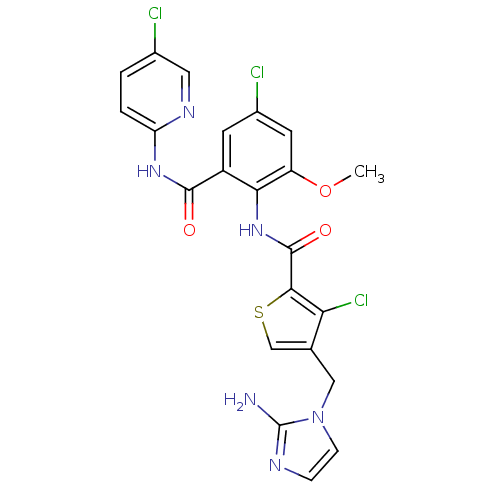 (4-[(2-amino-1H-imidazol-1-yl)methyl]-3-chloro-N-{4...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.00600 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17111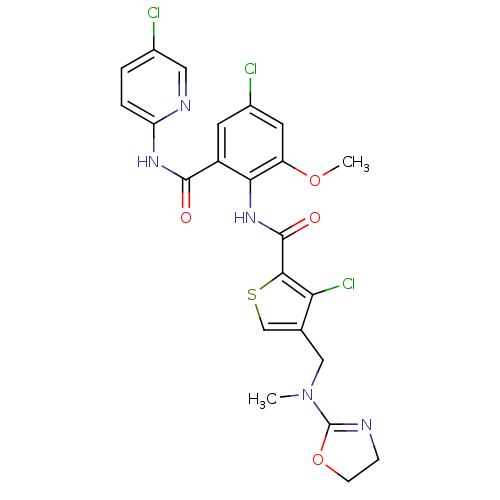 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid PDB UniChem Similars | PDB Article PubMed | 0.00700 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Muscarinic acetylcholine receptor M4 (Homo sapiens (Human)) | BDBM50296345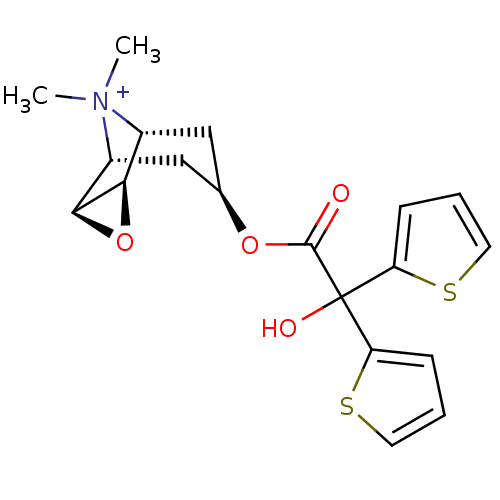 ((1R,2R,4S,5R,7S)-7-(2-hydroxy-2,2-di-thiophen-2-yl...) | PDB Reactome pathway KEGG UniProtKB/SwissProt DrugBank antibodypedia GoogleScholar AffyNet | Purchase CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 0.00780 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Pfizer Inc. Curated by ChEMBL | Assay Description Displacement of [3H]-NMS from human recombinant M4 receptor expressed in CHO cells after 24 hrs by filter binding assay | J Med Chem 54: 6888-904 (2011) Article DOI: 10.1021/jm200884j BindingDB Entry DOI: 10.7270/Q2CZ37JW | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Serine protease 1 (Bos taurus (bovine)) | BDBM50124947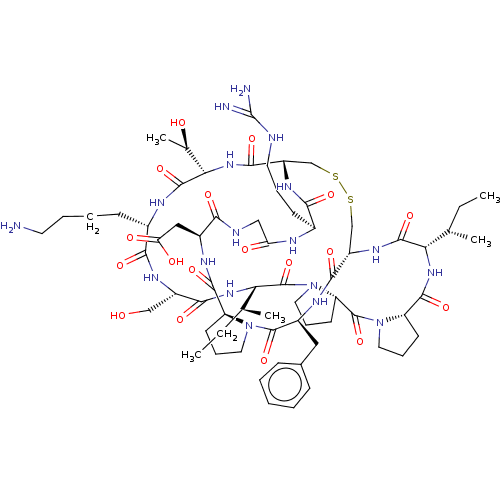 (CHEMBL453539) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL MCE PC cid PC sid UniChem Similars | Article PubMed | 0.00800 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The University of Queensland Curated by ChEMBL | Assay Description Inhibition of bovine beta-trypsin using Boc-QAR-MCA as substrate measured every 30 secs for 10 mins | Eur J Med Chem 155: 695-704 (2018) Article DOI: 10.1016/j.ejmech.2018.06.029 BindingDB Entry DOI: 10.7270/Q29026CK | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17137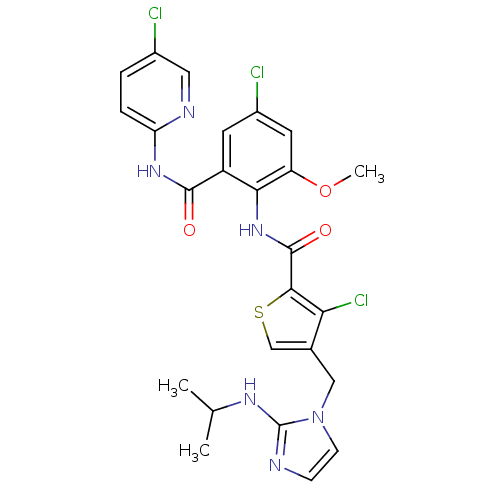 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 0.00800 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M3 (Homo sapiens (Human)) | BDBM50296345 ((1R,2R,4S,5R,7S)-7-(2-hydroxy-2,2-di-thiophen-2-yl...) | PDB Reactome pathway KEGG UniProtKB/SwissProt DrugBank antibodypedia GoogleScholar AffyNet | Purchase CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 0.00980 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Pfizer Inc. Curated by ChEMBL | Assay Description Displacement of [3H]-NMS from human recombinant M3 receptor expressed in CHO cells after 24 hrs by filter binding assay | J Med Chem 54: 6888-904 (2011) Article DOI: 10.1021/jm200884j BindingDB Entry DOI: 10.7270/Q2CZ37JW | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17130 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.0100 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Kappa-type opioid receptor (Cavia porcellus (domestic guinea pig)) | BDBM50242686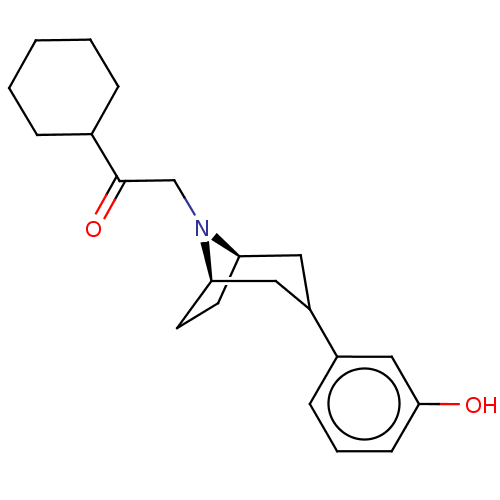 (CHEMBL4077003) | PDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 0.0100 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Theravance Biopharma US, Inc Curated by ChEMBL | Assay Description Displacement of [3H]DPN from guinea pig kappa opioid receptor expressed in CHOK1 cell membranes | Bioorg Med Chem Lett 27: 2926-2930 (2017) Article DOI: 10.1016/j.bmcl.2017.04.092 BindingDB Entry DOI: 10.7270/Q2WD430K | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Cathepsin D (Homo sapiens (Human)) | BDBM912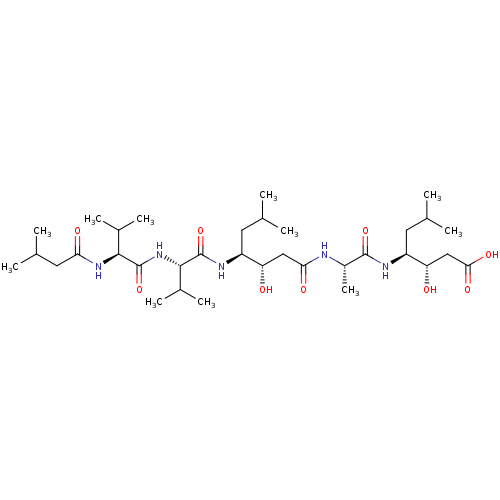 ((3S,4S)-3-hydroxy-4-[(2S)-2-[(3S,4S)-3-hydroxy-6-m...) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | Purchase CHEMBL MCE KEGG MMDB PC cid PC sid PDB UniChem Patents Similars | MMDB PubMed | 0.0100 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
University of Queensland Curated by ChEMBL | Assay Description Binding affinity against human cathepsin D | J Med Chem 43: 305-41 (2000) Checked by Author BindingDB Entry DOI: 10.7270/Q2JD4XH4 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17133 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.0100 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17131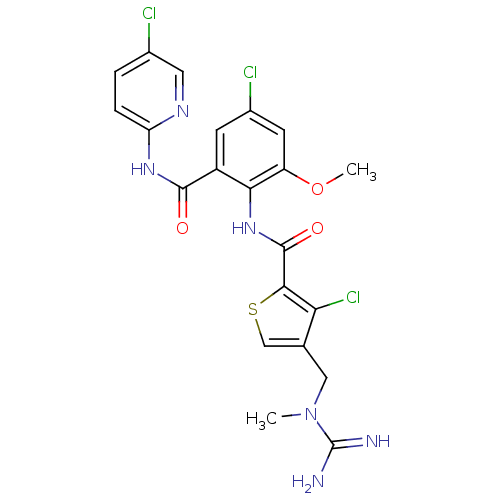 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.0100 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17123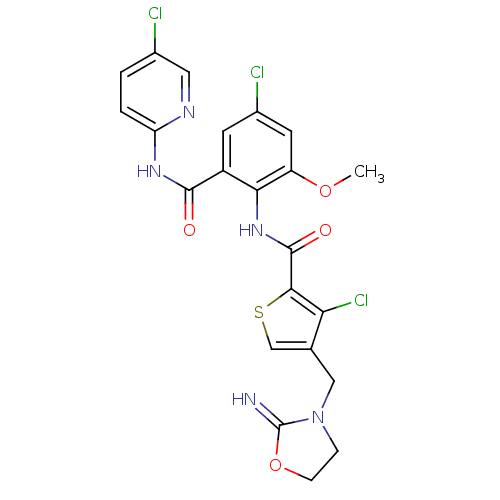 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.0100 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17124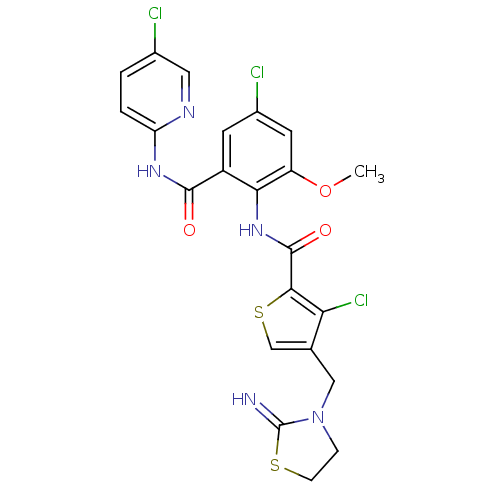 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.0100 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17125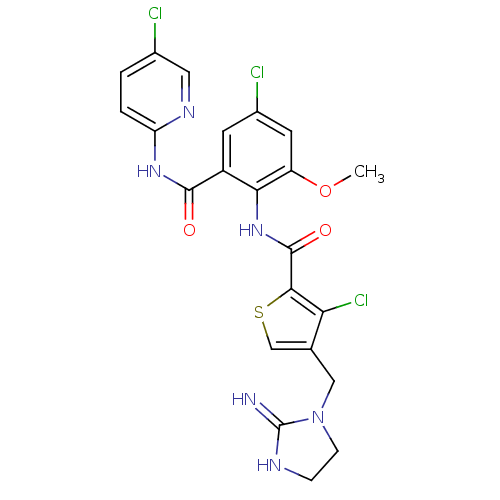 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.0100 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17128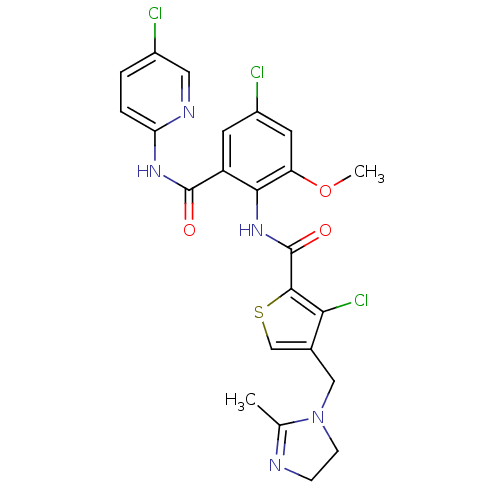 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.0100 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Kappa-type opioid receptor (Cavia porcellus (domestic guinea pig)) | BDBM50242686 (CHEMBL4077003) | PDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 0.0100 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Theravance Biopharma US, Inc Curated by ChEMBL | Assay Description Displacement of [3H]DPN from guinea pig kappa opioid receptor expressed in CHOK1 cell membranes | Bioorg Med Chem Lett 27: 2926-2930 (2017) Article DOI: 10.1016/j.bmcl.2017.04.092 BindingDB Entry DOI: 10.7270/Q2WD430K | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17112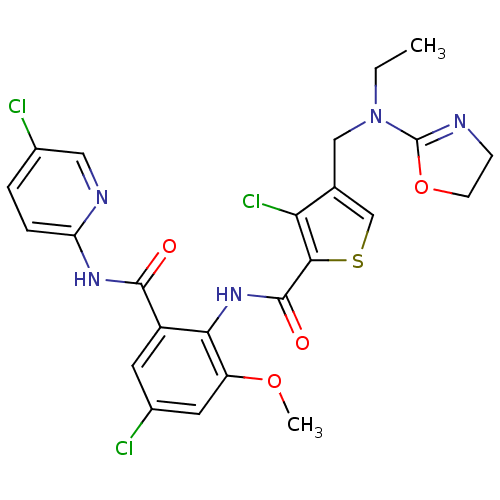 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.0120 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17118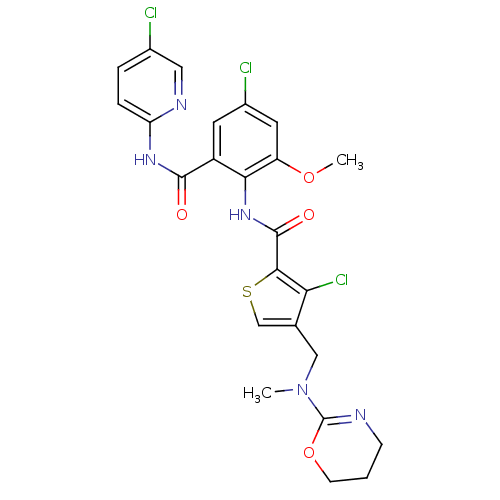 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 0.0120 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M1 (Homo sapiens (Human)) | BDBM50296345 ((1R,2R,4S,5R,7S)-7-(2-hydroxy-2,2-di-thiophen-2-yl...) | UniProtKB/SwissProt antibodypedia GoogleScholar AffyNet | Purchase CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 0.0130 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Pfizer Inc. Curated by ChEMBL | Assay Description Displacement of [3H]-NMS from human recombinant M1 receptor expressed in CHO cells after 24 hrs by filter binding assay | J Med Chem 54: 6888-904 (2011) Article DOI: 10.1021/jm200884j BindingDB Entry DOI: 10.7270/Q2CZ37JW | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Neutrophil elastase (Homo sapiens (Human)) | BDBM50036477 (2,6-Dichloro-3-(2-morpholin-4-yl-ethoxy)-benzoic a...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | 0.0130 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
University of Queensland Curated by ChEMBL | Assay Description Binding affinity against Elastase. | J Med Chem 43: 305-41 (2000) Checked by Author BindingDB Entry DOI: 10.7270/Q2JD4XH4 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17104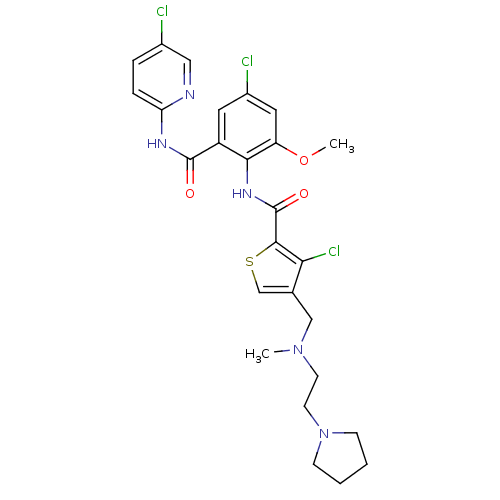 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.0190 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17138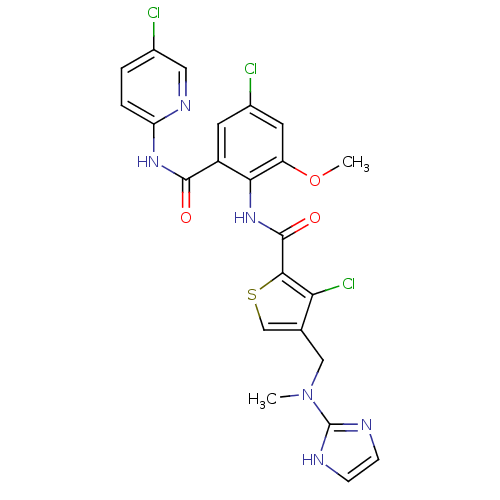 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 0.0200 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17107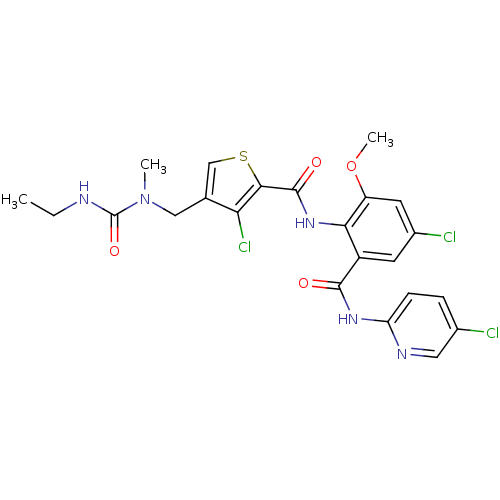 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 0.0200 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M2 (Homo sapiens (Human)) | BDBM50296345 ((1R,2R,4S,5R,7S)-7-(2-hydroxy-2,2-di-thiophen-2-yl...) | PDB UniProtKB/SwissProt antibodypedia GoogleScholar AffyNet | Purchase CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 0.0215 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Pfizer Inc. Curated by ChEMBL | Assay Description Displacement of [3H]-NMS from human recombinant M2 receptor expressed in CHO cells after 24 hrs by filter binding assay | J Med Chem 54: 6888-904 (2011) Article DOI: 10.1021/jm200884j BindingDB Entry DOI: 10.7270/Q2CZ37JW | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17110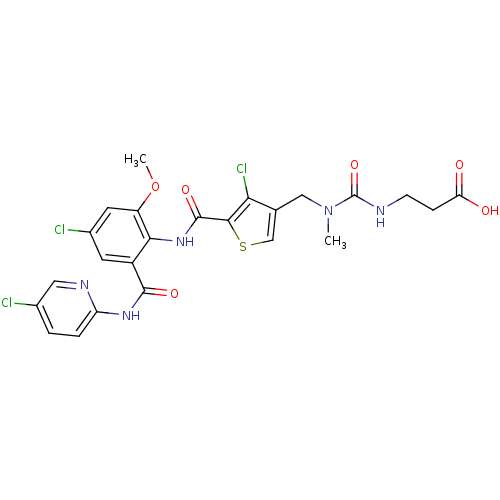 (3-[({[4-chloro-5-({4-chloro-2-[(5-chloropyridin-2-...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 0.0220 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17132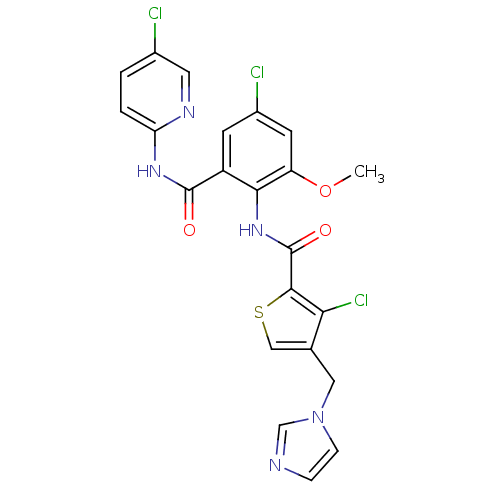 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.0220 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17120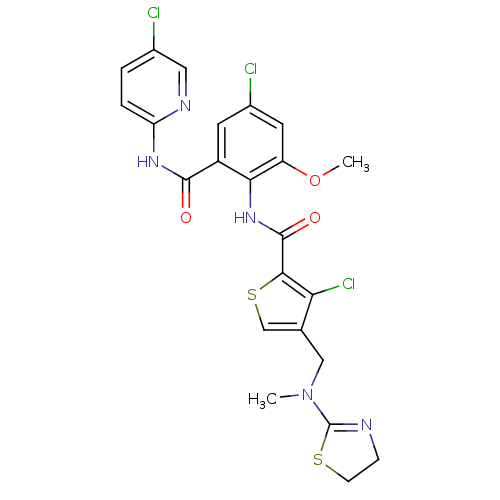 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.0240 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17103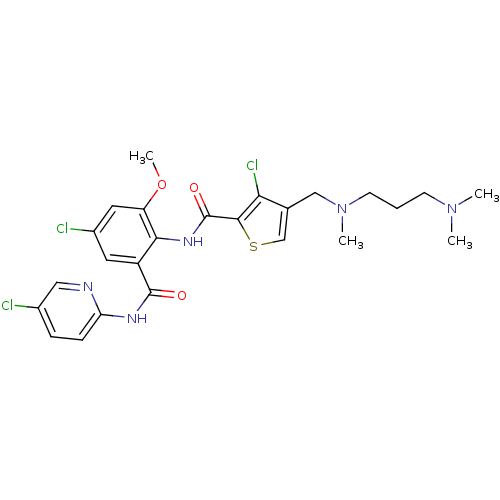 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.0240 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17119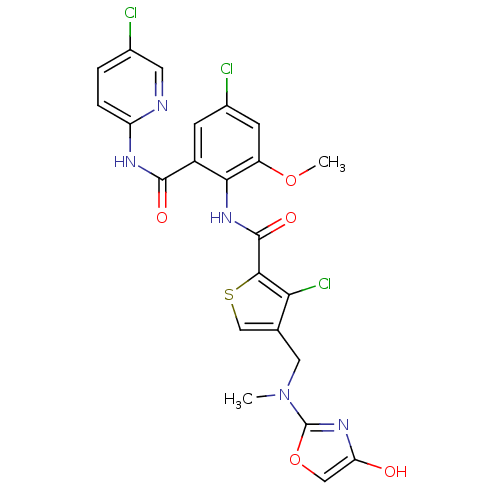 (3-chloro-N-{4-chloro-2-[(5-chloropyridin-2-yl)carb...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 0.0260 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM50088963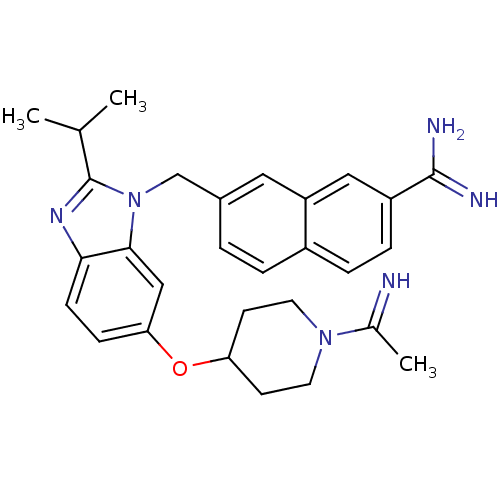 (7-{6-[1-(1-Imino-ethyl)-piperidin-4-yloxy]-2-isopr...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents Similars | PubMed | 0.0260 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences Curated by ChEMBL | Assay Description In vitro inhibitory activity against human Coagulation factor X | Bioorg Med Chem Lett 10: 963-6 (2000) BindingDB Entry DOI: 10.7270/Q2GQ6X0N | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Coagulation factor X (Homo sapiens (Human)) | BDBM17106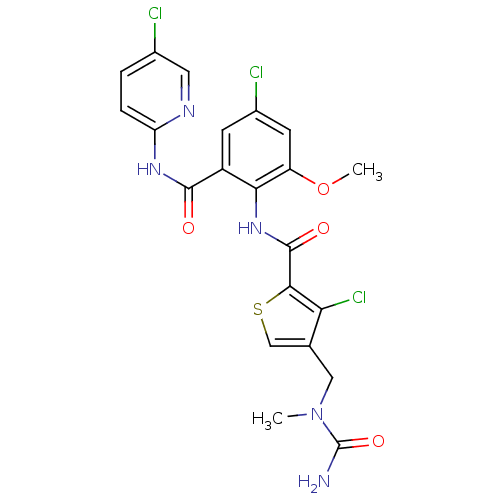 (4-{[carbamoyl(methyl)amino]methyl}-3-chloro-N-{4-c...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 0.0260 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Berlex Biosciences | Assay Description The enzyme activities were determined kinetically as the initial rate of cleavage of a peptide p-nitroanilide. Km for enzyme and substrate was determ... | J Med Chem 50: 2967-80 (2007) Article DOI: 10.1021/jm070125f BindingDB Entry DOI: 10.7270/Q2J101F5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Muscarinic acetylcholine receptor M5 (Homo sapiens (Human)) | BDBM50296345 ((1R,2R,4S,5R,7S)-7-(2-hydroxy-2,2-di-thiophen-2-yl...) | Reactome pathway KEGG UniProtKB/SwissProt DrugBank antibodypedia GoogleScholar AffyNet | Purchase CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 0.0266 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Pfizer Inc. Curated by ChEMBL | Assay Description Displacement of [3H]-NMS from human recombinant M5 receptor expressed in CHO cells after 24 hrs by filter binding assay | J Med Chem 54: 6888-904 (2011) Article DOI: 10.1021/jm200884j BindingDB Entry DOI: 10.7270/Q2CZ37JW | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [489-587] (Human immunodeficiency virus type 1) | BDBM430 (3-[(2-tert-butyl-4-hydroxy-5-methylphenyl)sulfanyl...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 0.0300 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Parke-Davis Pharmaceutical Research | Assay Description Sensitivity of HIV-1 protease activity to protease inhibitors was determined by a peptide substrate cleavage assay. Protease products were analyzed o... | J Med Chem 43: 843-58 (2000) Article DOI: 10.1021/jm990281p BindingDB Entry DOI: 10.7270/Q21N7Z9R | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Gag-Pol polyprotein [489-587] (Human immunodeficiency virus type 1) | BDBM50409174 (CHEMBL169119) | PDB MMDB UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 0.0300 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Parke-Davis Pharmaceutical Research | Assay Description Tested for binding affinity against HIV protease | J Med Chem 43: 843-58 (2000) Article DOI: 10.1021/jm990281p BindingDB Entry DOI: 10.7270/Q21N7Z9R | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [489-587] (Human immunodeficiency virus type 1) | BDBM472 (5-tert-butyl-4-{[(6S)-4-hydroxy-6-[2-(4-hydroxyphe...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 0.0300 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Parke-Davis Pharmaceutical Research | Assay Description Sensitivity of HIV-1 protease activity to protease inhibitors was determined by a peptide substrate cleavage assay. Protease products were analyzed o... | J Med Chem 43: 843-58 (2000) Article DOI: 10.1021/jm990281p BindingDB Entry DOI: 10.7270/Q21N7Z9R | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Gag-Pol polyprotein [489-587] (Human immunodeficiency virus type 1) | BDBM472 (5-tert-butyl-4-{[(6S)-4-hydroxy-6-[2-(4-hydroxyphe...) | PDB MMDB UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 0.0300 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Parke-Davis Pharmaceutical Research | Assay Description Tested for binding affinity against HIV protease | J Med Chem 43: 843-58 (2000) Article DOI: 10.1021/jm990281p BindingDB Entry DOI: 10.7270/Q21N7Z9R | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Serine protease 1 (Homo sapiens (Human)) | BDBM50124947 (CHEMBL453539) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL MCE PC cid PC sid UniChem Similars | Article PubMed | 0.0300 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The University of Queensland Curated by ChEMBL | Assay Description Inhibition of human cationic trypsin using Boc-VPR-MCA as substrate measured every 30 secs for 10 mins | Eur J Med Chem 155: 695-704 (2018) Article DOI: 10.1016/j.ejmech.2018.06.029 BindingDB Entry DOI: 10.7270/Q29026CK | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Melatonin receptor type 1B (Homo sapiens (Human)) | BDBM50425203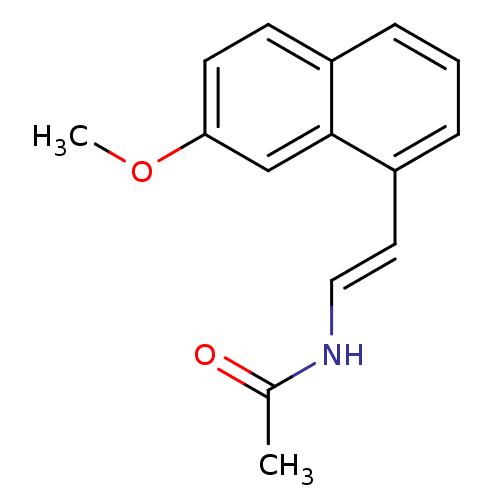 (CHEMBL2314255) | PDB Reactome pathway KEGG UniProtKB/SwissProt DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 0.0300 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Universit£ de Paris-Sud Curated by ChEMBL | Assay Description Displacement of [125I]2-iodomelatonin from human recombinant MT2 receptor expressed in HEK293 cells after 2 hrs by gamma counting | Bioorg Med Chem Lett 23: 430-4 (2012) Article DOI: 10.1016/j.bmcl.2012.11.069 BindingDB Entry DOI: 10.7270/Q2QJ7JM0 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Melatonin receptor type 1B (Homo sapiens (Human)) | BDBM50230960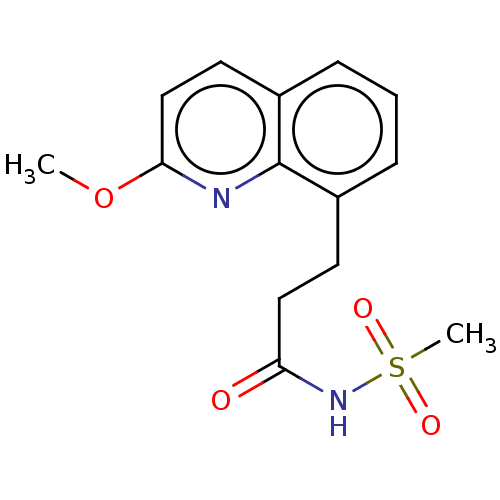 (CHEMBL4089965) | PDB Reactome pathway KEGG UniProtKB/SwissProt DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 0.0300 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Universit£ de Lille Curated by ChEMBL | Assay Description Displacement of 2-[125I]iodomelatonin from human MT2 receptor expressed in HEK or CHO cell membranes after 120 mins | Eur J Med Chem 127: 621-631 (2017) Article DOI: 10.1016/j.ejmech.2016.12.013 BindingDB Entry DOI: 10.7270/Q24F1SZS | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Melatonin receptor type 1A (Homo sapiens (Human)) | BDBM50425202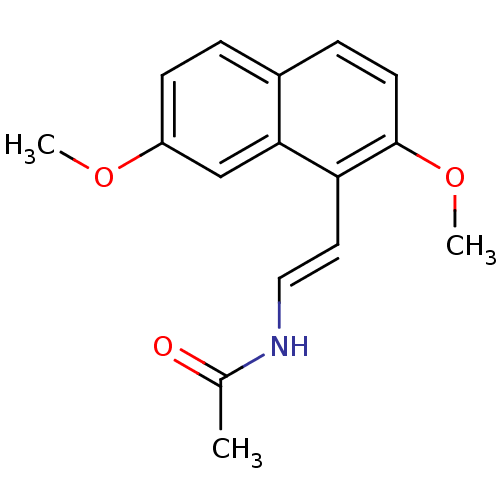 (CHEMBL2314259) | PDB Reactome pathway KEGG UniProtKB/SwissProt DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 0.0300 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Universit£ de Paris-Sud Curated by ChEMBL | Assay Description Displacement of [125I]2-iodomelatonin from human recombinant MT1 receptor expressed in HEK293 cells after 2 hrs by gamma counting | Bioorg Med Chem Lett 23: 430-4 (2012) Article DOI: 10.1016/j.bmcl.2012.11.069 BindingDB Entry DOI: 10.7270/Q2QJ7JM0 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Melatonin receptor type 1A (Homo sapiens (Human)) | BDBM50117886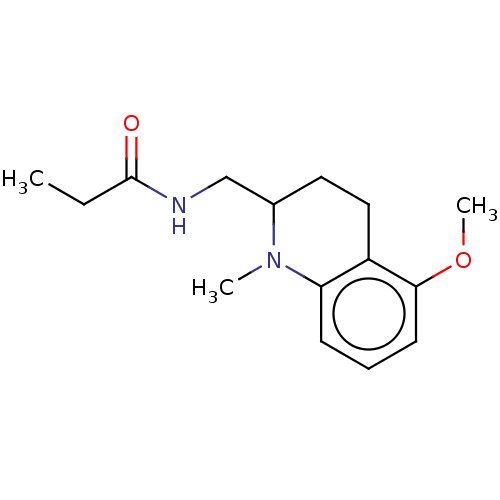 (CHEMBL3612603) | PDB Reactome pathway KEGG UniProtKB/SwissProt DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents | Article PubMed | 0.0300 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Universit£ degli Studi di Urbino"Carlo Bo" Curated by ChEMBL | Assay Description Displacement of 2-[125I]-Iodomelatonin from human MT1 receptor transfected in CHO cell membranes after 120 mins | J Med Chem 58: 7512-25 (2015) Article DOI: 10.1021/acs.jmedchem.5b01066 BindingDB Entry DOI: 10.7270/Q20R9R6D | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Trypsin (Homo sapiens (Human)) | BDBM50125046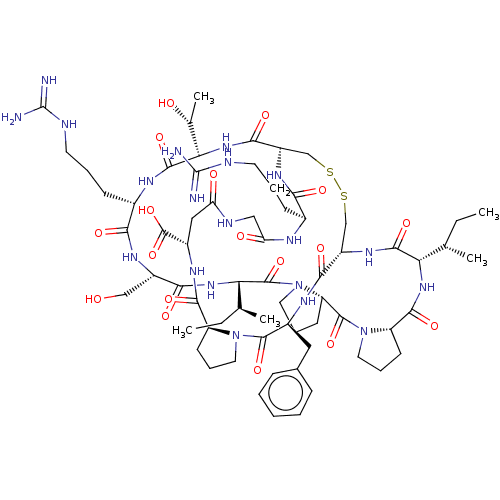 (CHEMBL3623792) | PDB KEGG UniProtKB/SwissProt antibodypedia GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem | Article PubMed | 0.0300 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Queensland University of Technology Curated by ChEMBL | Assay Description Inhibition of beta-trypsin (unknown origin) using Bz-FVRpNA substrate by spectrophotometry method | J Med Chem 58: 8257-68 (2015) Article DOI: 10.1021/acs.jmedchem.5b01148 BindingDB Entry DOI: 10.7270/Q2PR7XSJ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Displayed 1 to 50 (of 39794 total ) | Next | Last >> |