

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DNA polymerase III PolC-type (Bacillus subtilis) | BDBM50182669 ((S)-3-{4-[1-(1-cyclopropyl-3-carboxy-4-oxo-6,8-dif...) | KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
GLSynthesis Inc. Curated by ChEMBL | Assay Description Binding affinity to Bacillus subtilis DNA polymerase3C | J Med Chem 49: 1455-65 (2006) Article DOI: 10.1021/jm0510023 BindingDB Entry DOI: 10.7270/Q23J3DR1 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA polymerase III PolC-type (Bacillus subtilis) | BDBM50182665 ((R)-3-{4-[1-(1-cyclopropyl-3-carboxy-4-oxo-6,8-dif...) | KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 7 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
GLSynthesis Inc. Curated by ChEMBL | Assay Description Binding affinity to Bacillus subtilis DNA polymerase3C | J Med Chem 49: 1455-65 (2006) Article DOI: 10.1021/jm0510023 BindingDB Entry DOI: 10.7270/Q23J3DR1 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA polymerase III PolC-type (Bacillus subtilis) | BDBM50182666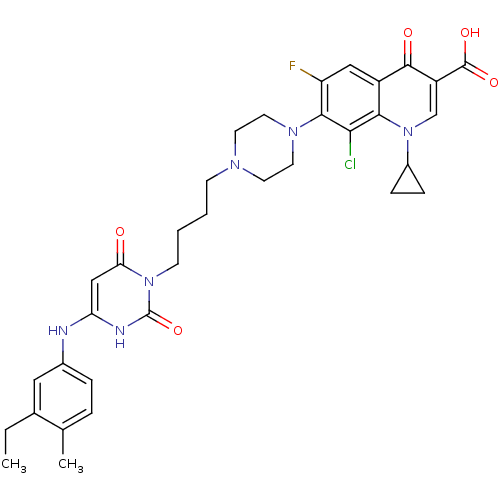 (3-{4-[1-(1-cyclopropyl-3-carboxy-4-oxo-6-fluoro-8-...) | KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 10 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
GLSynthesis Inc. Curated by ChEMBL | Assay Description Binding affinity to Bacillus subtilis DNA polymerase3C | J Med Chem 49: 1455-65 (2006) Article DOI: 10.1021/jm0510023 BindingDB Entry DOI: 10.7270/Q23J3DR1 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA polymerase III PolC-type (Bacillus subtilis) | BDBM50182667 (3-{4-[1-(1-cyclopropyl-3-carboxy-4-oxo-6,8-difluor...) | KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents Similars | Article PubMed | 11 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
GLSynthesis Inc. Curated by ChEMBL | Assay Description Binding affinity to Bacillus subtilis DNA polymerase3C | J Med Chem 49: 1455-65 (2006) Article DOI: 10.1021/jm0510023 BindingDB Entry DOI: 10.7270/Q23J3DR1 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA polymerase III PolC-type (Bacillus subtilis) | BDBM50182662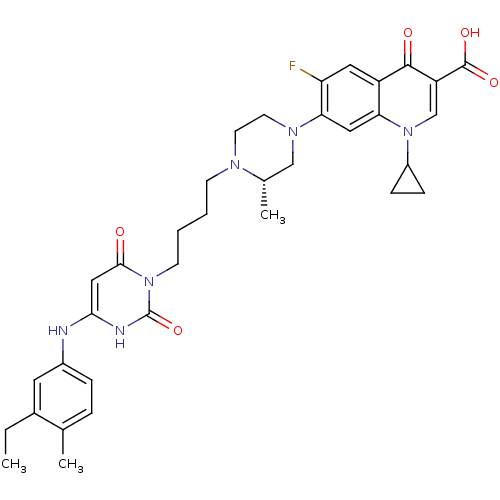 ((S)-3-{4-[1-(1-cyclopropyl-3-carboxy-4-oxo-6-fluor...) | KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents Similars | Article PubMed | 11 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
GLSynthesis Inc. Curated by ChEMBL | Assay Description Binding affinity to Bacillus subtilis DNA polymerase3C | J Med Chem 49: 1455-65 (2006) Article DOI: 10.1021/jm0510023 BindingDB Entry DOI: 10.7270/Q23J3DR1 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA polymerase III PolC-type (Bacillus subtilis) | BDBM50182674 ((R)-3-{4-[1-(1-cyclopropyl-3-carboxy-4-oxo-6-fluor...) | KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents Similars | Article PubMed | 12 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
GLSynthesis Inc. Curated by ChEMBL | Assay Description Binding affinity to Bacillus subtilis DNA polymerase3C | J Med Chem 49: 1455-65 (2006) Article DOI: 10.1021/jm0510023 BindingDB Entry DOI: 10.7270/Q23J3DR1 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA polymerase III PolC-type (Bacillus subtilis) | BDBM50182659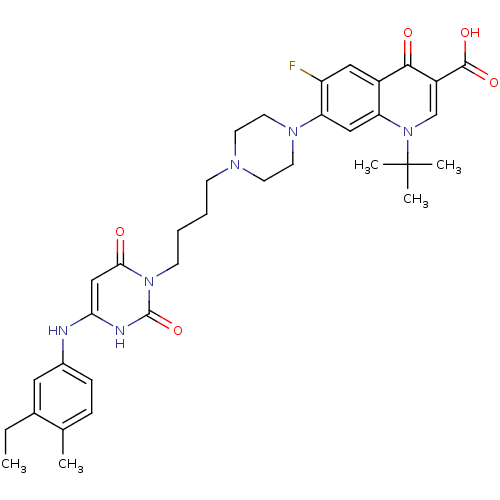 (3-{4-[1-(1-tert-butyl-3-carboxy-4-oxo-6-fluoro-7-q...) | KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 13 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
GLSynthesis Inc. Curated by ChEMBL | Assay Description Binding affinity to Bacillus subtilis DNA polymerase3C | J Med Chem 49: 1455-65 (2006) Article DOI: 10.1021/jm0510023 BindingDB Entry DOI: 10.7270/Q23J3DR1 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA polymerase III PolC-type (Bacillus subtilis) | BDBM50182663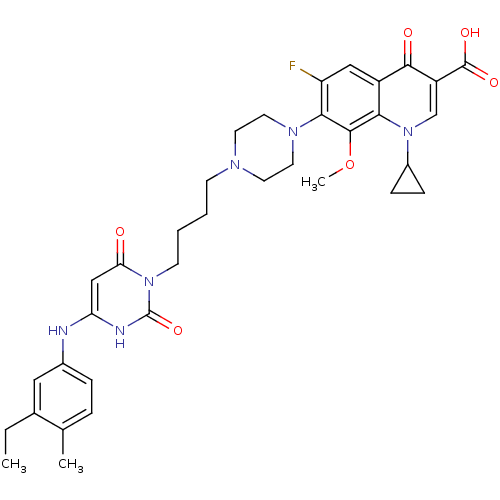 (3-{4-[1-(1-cyclopropyl-3-carboxy-4-oxo-6-fluoro-8-...) | KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 14 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
GLSynthesis Inc. Curated by ChEMBL | Assay Description Binding affinity to Bacillus subtilis DNA polymerase3C | J Med Chem 49: 1455-65 (2006) Article DOI: 10.1021/jm0510023 BindingDB Entry DOI: 10.7270/Q23J3DR1 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA polymerase III PolC-type (Bacillus subtilis) | BDBM50182673 (3-{4-[1-(1-(4-fluorophenyl)-3-carboxy-4-oxo-6-fluo...) | KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 17 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
GLSynthesis Inc. Curated by ChEMBL | Assay Description Binding affinity to Bacillus subtilis DNA polymerase3C | J Med Chem 49: 1455-65 (2006) Article DOI: 10.1021/jm0510023 BindingDB Entry DOI: 10.7270/Q23J3DR1 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA polymerase III PolC-type (Bacillus subtilis) | BDBM50182676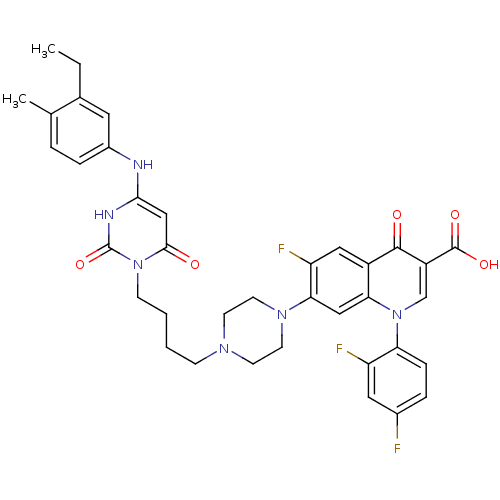 (3-{4-[1-(1-{2,4-difluorophenyl}-3-carboxy-4-oxo-6-...) | KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents Similars | Article PubMed | 18 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
GLSynthesis Inc. Curated by ChEMBL | Assay Description Binding affinity to Bacillus subtilis DNA polymerase3C | J Med Chem 49: 1455-65 (2006) Article DOI: 10.1021/jm0510023 BindingDB Entry DOI: 10.7270/Q23J3DR1 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA polymerase III PolC-type (Bacillus subtilis) | BDBM50182672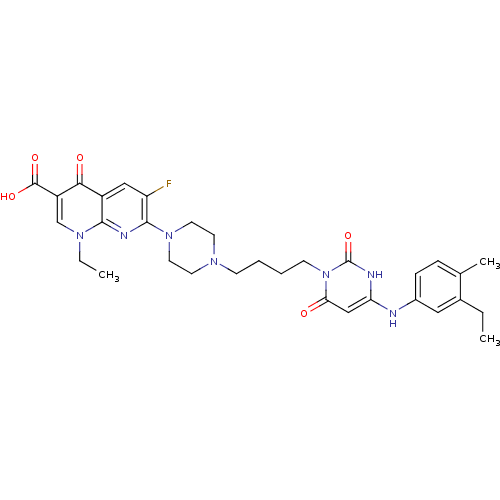 (1-ethyl-7-(4-(4-(4-(3-ethyl-4-methylphenylamino)-2...) | KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents Similars | Article PubMed | 18 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
GLSynthesis Inc. Curated by ChEMBL | Assay Description Binding affinity to Bacillus subtilis DNA polymerase3C | J Med Chem 49: 1455-65 (2006) Article DOI: 10.1021/jm0510023 BindingDB Entry DOI: 10.7270/Q23J3DR1 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA polymerase III PolC-type (Bacillus subtilis) | BDBM50182671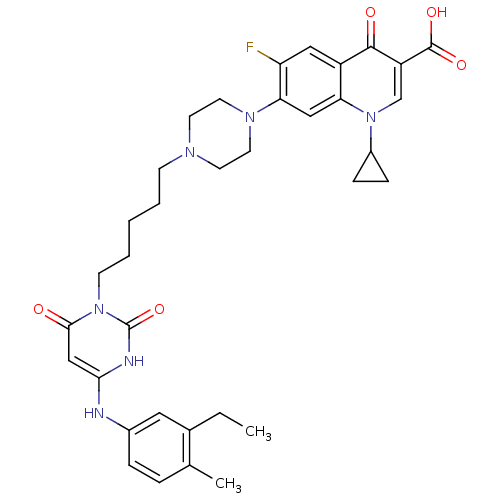 (3-{5-[1-(1-cyclopropyl-3-carboxy-4-oxo-6-fluoro-7-...) | KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 19 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
GLSynthesis Inc. Curated by ChEMBL | Assay Description Binding affinity to Bacillus subtilis DNA polymerase3C | J Med Chem 49: 1455-65 (2006) Article DOI: 10.1021/jm0510023 BindingDB Entry DOI: 10.7270/Q23J3DR1 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA polymerase III PolC-type (Bacillus subtilis) | BDBM21688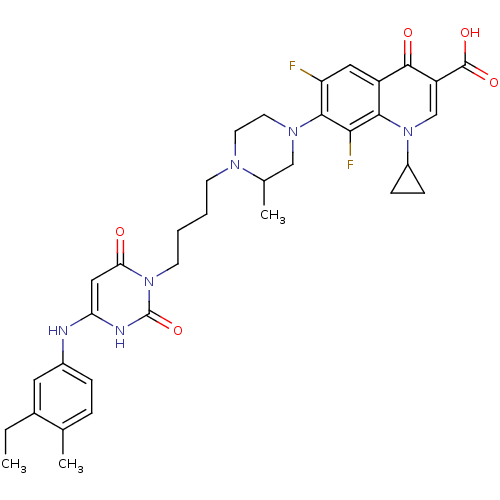 (1-cyclopropyl-7-[4-(4-{4-[(3-ethyl-4-methylphenyl)...) | KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents Similars | Article PubMed | 19 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
GLSynthesis Inc. Curated by ChEMBL | Assay Description Binding affinity to Bacillus subtilis DNA polymerase3C | J Med Chem 49: 1455-65 (2006) Article DOI: 10.1021/jm0510023 BindingDB Entry DOI: 10.7270/Q23J3DR1 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA polymerase III PolC-type (Bacillus subtilis) | BDBM50182670 (3-{4-[1-(1-ethyl-3-carboxy-4-oxo-6,8-difluoro-7-qu...) | KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents Similars | Article PubMed | 21 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
GLSynthesis Inc. Curated by ChEMBL | Assay Description Binding affinity to Bacillus subtilis DNA polymerase3C | J Med Chem 49: 1455-65 (2006) Article DOI: 10.1021/jm0510023 BindingDB Entry DOI: 10.7270/Q23J3DR1 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA polymerase III PolC-type (Bacillus subtilis) | BDBM50182668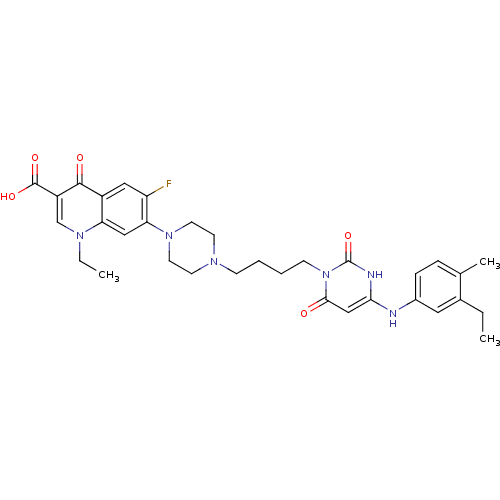 (3-{4-[1-(1-ethyl-3-carboxy-4-oxo-6-fluoro-7-quinol...) | KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents Similars | Article PubMed | 24 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
GLSynthesis Inc. Curated by ChEMBL | Assay Description Binding affinity to Bacillus subtilis DNA polymerase3C | J Med Chem 49: 1455-65 (2006) Article DOI: 10.1021/jm0510023 BindingDB Entry DOI: 10.7270/Q23J3DR1 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA polymerase III PolC-type (Bacillus subtilis) | BDBM50182675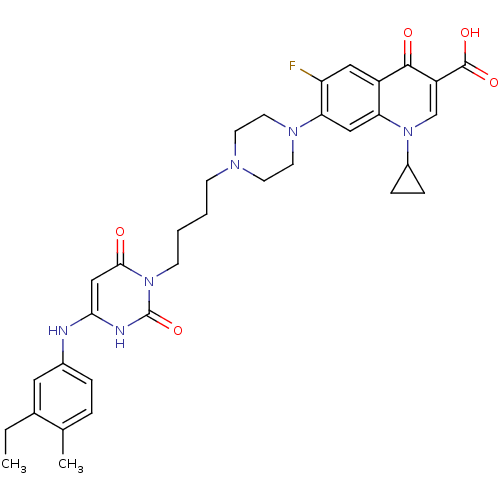 (3-{4-[1-(1-cyclopropyl-3-carboxy-4-oxo-6-fluoro-7-...) | KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents Similars | Article PubMed | 24 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
GLSynthesis Inc. Curated by ChEMBL | Assay Description Binding affinity to Bacillus subtilis DNA polymerase3C | J Med Chem 49: 1455-65 (2006) Article DOI: 10.1021/jm0510023 BindingDB Entry DOI: 10.7270/Q23J3DR1 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA polymerase III PolC-type (Bacillus subtilis) | BDBM50182656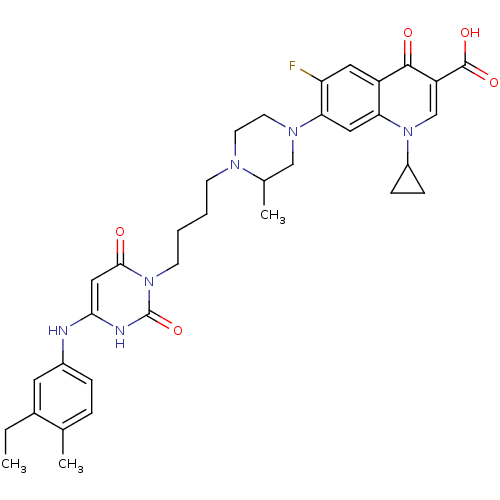 (3-{4-[1-(1-cyclopropyl-3-carboxy-4-oxo-6-fluoro-7-...) | KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 26 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
GLSynthesis Inc. Curated by ChEMBL | Assay Description Binding affinity to Bacillus subtilis DNA polymerase3C | J Med Chem 49: 1455-65 (2006) Article DOI: 10.1021/jm0510023 BindingDB Entry DOI: 10.7270/Q23J3DR1 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA polymerase III PolC-type (Bacillus subtilis) | BDBM50182657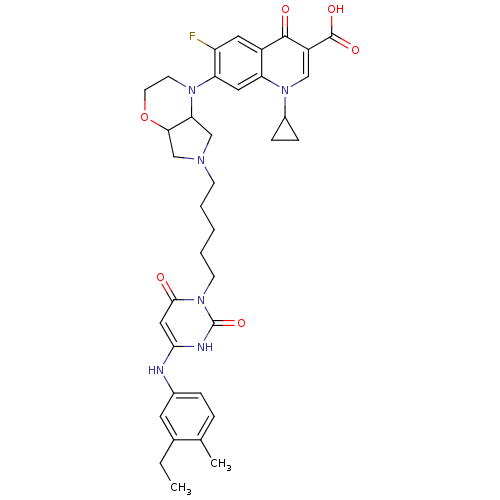 (3-{5-[1-(1-cyclopropyl-3-carboxy-4-oxo-6-fluoro-7-...) | KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 33 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
GLSynthesis Inc. Curated by ChEMBL | Assay Description Binding affinity to Bacillus subtilis DNA polymerase3C | J Med Chem 49: 1455-65 (2006) Article DOI: 10.1021/jm0510023 BindingDB Entry DOI: 10.7270/Q23J3DR1 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA polymerase III PolC-type (Bacillus subtilis) | BDBM50182664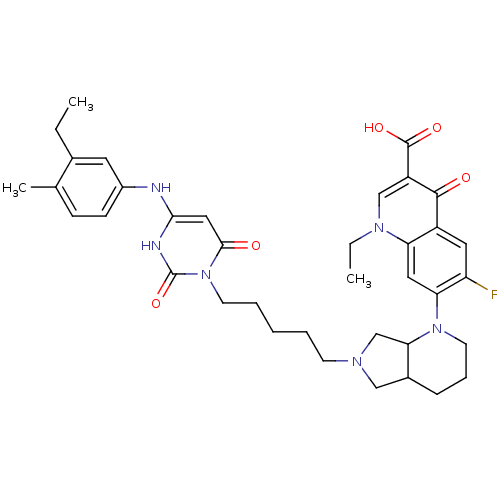 (3-{5-[1-(1-ethyl-3-carboxy-4-oxo-6-fluoro-7-quinol...) | KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 37 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
GLSynthesis Inc. Curated by ChEMBL | Assay Description Binding affinity to Bacillus subtilis DNA polymerase3C | J Med Chem 49: 1455-65 (2006) Article DOI: 10.1021/jm0510023 BindingDB Entry DOI: 10.7270/Q23J3DR1 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA polymerase III PolC-type (Bacillus subtilis) | BDBM50182658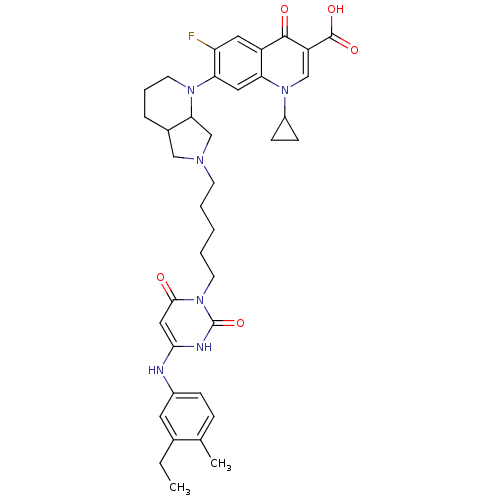 (3-{5-[1-(1-cyclopropyl-3-carboxy-6-fluoro-4-oxo-7-...) | KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 41 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
GLSynthesis Inc. Curated by ChEMBL | Assay Description Binding affinity to Bacillus subtilis DNA polymerase3C | J Med Chem 49: 1455-65 (2006) Article DOI: 10.1021/jm0510023 BindingDB Entry DOI: 10.7270/Q23J3DR1 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA polymerase III PolC-type (Bacillus subtilis) | BDBM21686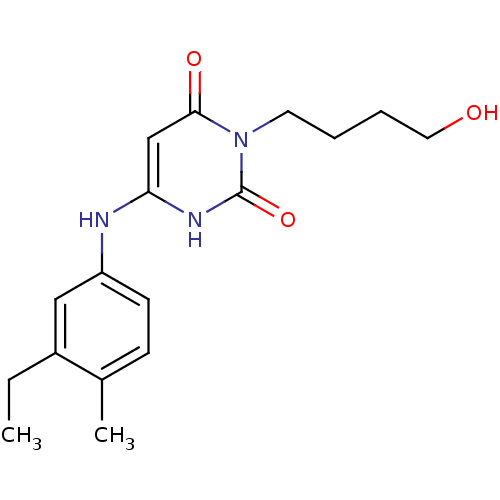 (3-(4-hydroxybutyl)-6-(3-ethyl-4-methylanilino)urac...) | KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Patents Similars | Article PubMed | 66 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
GLSynthesis Inc. Curated by ChEMBL | Assay Description Binding affinity to Bacillus subtilis DNA polymerase3C | J Med Chem 49: 1455-65 (2006) Article DOI: 10.1021/jm0510023 BindingDB Entry DOI: 10.7270/Q23J3DR1 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA polymerase III PolC-type (Bacillus subtilis) | BDBM50182660 (3-{4-[1-(1-cyclopropyl-3-carboxy-4-oxo-6-fluoro-7-...) | KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 95 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
GLSynthesis Inc. Curated by ChEMBL | Assay Description Binding affinity to Bacillus subtilis DNA polymerase3C | J Med Chem 49: 1455-65 (2006) Article DOI: 10.1021/jm0510023 BindingDB Entry DOI: 10.7270/Q23J3DR1 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA polymerase III PolC-type (Bacillus subtilis) | BDBM50182661 (3-{4-[1-(1-cyclopropyl-3-carboxy-4-oxo-6-fluoro-7-...) | KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 2.90E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
GLSynthesis Inc. Curated by ChEMBL | Assay Description Binding affinity to Bacillus subtilis DNA polymerase3C | J Med Chem 49: 1455-65 (2006) Article DOI: 10.1021/jm0510023 BindingDB Entry DOI: 10.7270/Q23J3DR1 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Genome polyprotein (Hepatitis C Virus) | BDBM50495931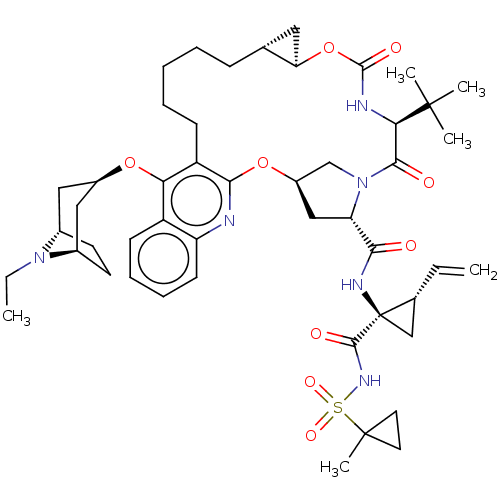 (CHEMBL3348818) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 0.0100 | n/a | n/a | n/a | n/a | n/a | n/a |
Merck Research Laboratories Curated by ChEMBL | Assay Description Inhibition of Hepatitis C virus genotype 1b NS3/4A protease R155K mutant assessed as substrate cleavage using Ac-C(Eu)DDMEEAbu(COO)ASK(QSY7)-amide as... | ACS Med Chem Lett 5: 264-9 (2014) Article DOI: 10.1021/ml400466p BindingDB Entry DOI: 10.7270/Q22R3VNT | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Genome polyprotein (Hepatitis C Virus) | BDBM50495935 (CHEMBL3120475) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 0.0200 | n/a | n/a | n/a | n/a | n/a | n/a |
Merck Research Laboratories Curated by ChEMBL | Assay Description Inhibition of Hepatitis C virus genotype 1b NS3/4A protease R155K mutant assessed as substrate cleavage using Ac-C(Eu)DDMEEAbu(COO)ASK(QSY7)-amide as... | ACS Med Chem Lett 5: 264-9 (2014) Article DOI: 10.1021/ml400466p BindingDB Entry DOI: 10.7270/Q22R3VNT | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Genome polyprotein (Hepatitis C Virus) | BDBM50495928 (CHEMBL3349199) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 0.0200 | n/a | n/a | n/a | n/a | n/a | n/a |
Merck Research Laboratories Curated by ChEMBL | Assay Description Inhibition of Hepatitis C virus genotype 1b NS3/4A protease R155K mutant assessed as substrate cleavage using Ac-C(Eu)DDMEEAbu(COO)ASK(QSY7)-amide as... | ACS Med Chem Lett 5: 264-9 (2014) Article DOI: 10.1021/ml400466p BindingDB Entry DOI: 10.7270/Q22R3VNT | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Genome polyprotein (Hepatitis C Virus) | BDBM50495927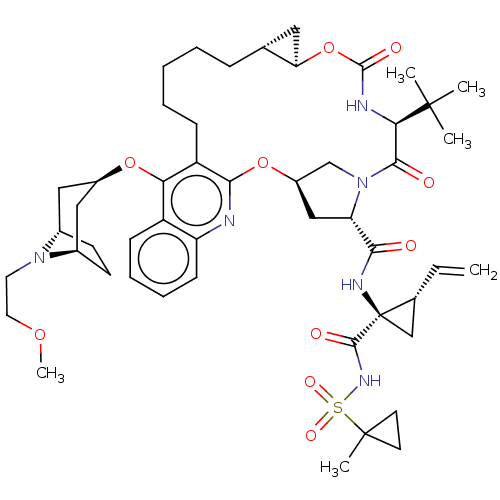 (CHEMBL3348817) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 0.0200 | n/a | n/a | n/a | n/a | n/a | n/a |
Merck Research Laboratories Curated by ChEMBL | Assay Description Inhibition of Hepatitis C virus genotype 1b NS3/4A protease R155K mutant assessed as substrate cleavage using Ac-C(Eu)DDMEEAbu(COO)ASK(QSY7)-amide as... | ACS Med Chem Lett 5: 264-9 (2014) Article DOI: 10.1021/ml400466p BindingDB Entry DOI: 10.7270/Q22R3VNT | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Genome polyprotein (Hepatitis C Virus) | BDBM50495924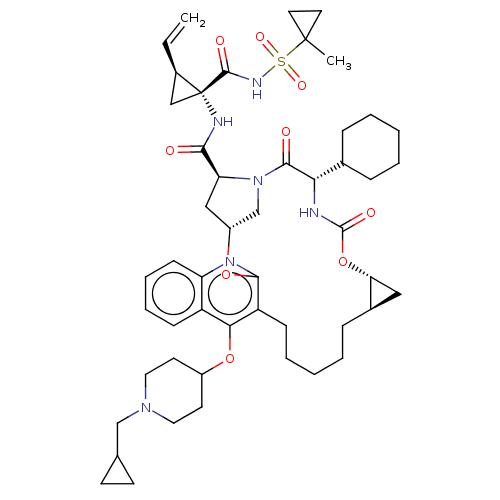 (CHEMBL3120477) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 0.0300 | n/a | n/a | n/a | n/a | n/a | n/a |
Merck Research Laboratories Curated by ChEMBL | Assay Description Inhibition of Hepatitis C virus genotype 1b NS3/4A protease R155K mutant assessed as substrate cleavage using Ac-C(Eu)DDMEEAbu(COO)ASK(QSY7)-amide as... | ACS Med Chem Lett 5: 264-9 (2014) Article DOI: 10.1021/ml400466p BindingDB Entry DOI: 10.7270/Q22R3VNT | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Genome polyprotein (Hepatitis C Virus) | BDBM50495931 (CHEMBL3348818) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 0.0400 | n/a | n/a | n/a | n/a | n/a | n/a |
Merck Research Laboratories Curated by ChEMBL | Assay Description Inhibition of Hepatitis C virus genotype 1b NS3/4A protease D168Y mutant assessed as substrate cleavage using Ac-C(Eu)DDMEEAbu(COO)ASK(QSY7)-amide as... | ACS Med Chem Lett 5: 264-9 (2014) Article DOI: 10.1021/ml400466p BindingDB Entry DOI: 10.7270/Q22R3VNT | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Genome polyprotein (Hepatitis C Virus) | BDBM50495936 (CHEMBL3120492) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 0.0500 | n/a | n/a | n/a | n/a | n/a | n/a |
Merck Research Laboratories Curated by ChEMBL | Assay Description Inhibition of Hepatitis C virus genotype 1b NS3/4A protease R155K mutant assessed as substrate cleavage using Ac-C(Eu)DDMEEAbu(COO)ASK(QSY7)-amide as... | ACS Med Chem Lett 5: 264-9 (2014) Article DOI: 10.1021/ml400466p BindingDB Entry DOI: 10.7270/Q22R3VNT | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Genome polyprotein (Hepatitis C Virus) | BDBM50495928 (CHEMBL3349199) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 0.0500 | n/a | n/a | n/a | n/a | n/a | n/a |
Merck Research Laboratories Curated by ChEMBL | Assay Description Inhibition of Hepatitis C virus genotype 1b NS3/4A protease D168Y mutant assessed as substrate cleavage using Ac-C(Eu)DDMEEAbu(COO)ASK(QSY7)-amide as... | ACS Med Chem Lett 5: 264-9 (2014) Article DOI: 10.1021/ml400466p BindingDB Entry DOI: 10.7270/Q22R3VNT | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Genome polyprotein (Hepatitis C Virus) | BDBM50495943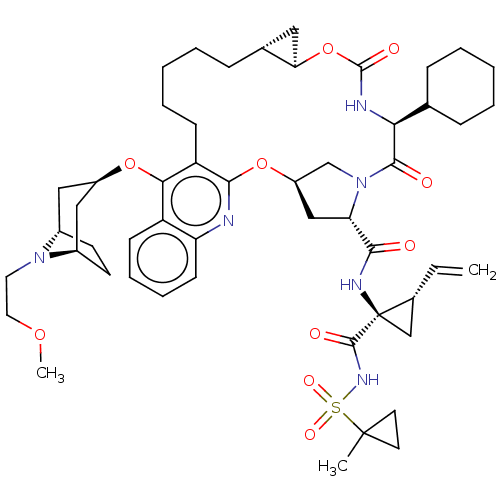 (CHEMBL3349202) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 0.0500 | n/a | n/a | n/a | n/a | n/a | n/a |
Merck Research Laboratories Curated by ChEMBL | Assay Description Inhibition of Hepatitis C virus genotype 1b NS3/4A protease R155K mutant assessed as substrate cleavage using Ac-C(Eu)DDMEEAbu(COO)ASK(QSY7)-amide as... | ACS Med Chem Lett 5: 264-9 (2014) Article DOI: 10.1021/ml400466p BindingDB Entry DOI: 10.7270/Q22R3VNT | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Genome polyprotein (Hepatitis C Virus) | BDBM50495927 (CHEMBL3348817) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 0.0600 | n/a | n/a | n/a | n/a | n/a | n/a |
Merck Research Laboratories Curated by ChEMBL | Assay Description Inhibition of Hepatitis C virus genotype 1b NS3/4A protease D168Y mutant assessed as substrate cleavage using Ac-C(Eu)DDMEEAbu(COO)ASK(QSY7)-amide as... | ACS Med Chem Lett 5: 264-9 (2014) Article DOI: 10.1021/ml400466p BindingDB Entry DOI: 10.7270/Q22R3VNT | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Genome polyprotein (Hepatitis C Virus) | BDBM50495929 (CHEMBL3120493) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 0.0600 | n/a | n/a | n/a | n/a | n/a | n/a |
Merck Research Laboratories Curated by ChEMBL | Assay Description Inhibition of Hepatitis C virus genotype 1b NS3/4A protease R155K mutant assessed as substrate cleavage using Ac-C(Eu)DDMEEAbu(COO)ASK(QSY7)-amide as... | ACS Med Chem Lett 5: 264-9 (2014) Article DOI: 10.1021/ml400466p BindingDB Entry DOI: 10.7270/Q22R3VNT | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Genome polyprotein (Hepatitis C Virus) | BDBM50485492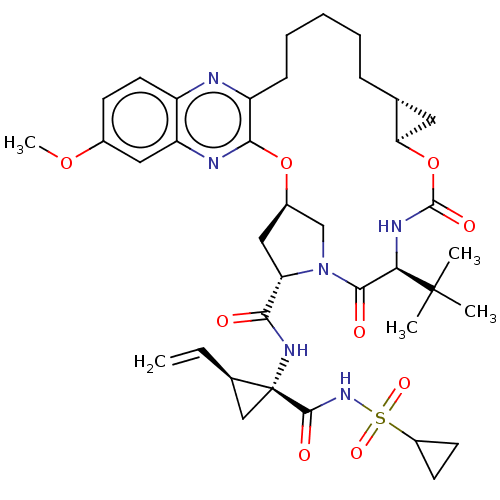 (Grazoprevir | Grazoprevir monohydrate | MK-5172 | ...) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | Purchase MCE PC cid PC sid PDB UniChem | PDB Article PubMed | n/a | n/a | 0.0700 | n/a | n/a | n/a | n/a | n/a | n/a |
Merck Research Laboratories Curated by ChEMBL | Assay Description Inhibition of Hepatitis C virus genotype 1b NS3/4A protease R155K mutant assessed as substrate cleavage using Ac-C(Eu)DDMEEAbu(COO)ASK(QSY7)-amide as... | ACS Med Chem Lett 5: 264-9 (2014) Article DOI: 10.1021/ml400466p BindingDB Entry DOI: 10.7270/Q22R3VNT | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Genome polyprotein (Hepatitis C Virus) | BDBM50495928 (CHEMBL3349199) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 0.0800 | n/a | n/a | n/a | n/a | n/a | n/a |
Merck Research Laboratories Curated by ChEMBL | Assay Description Inhibition of Hepatitis C virus genotype 1b NS3/4A protease A156T mutant assessed as substrate cleavage using Ac-C(Eu)DDMEEAbu(COO)ASK(QSY7)-amide as... | ACS Med Chem Lett 5: 264-9 (2014) Article DOI: 10.1021/ml400466p BindingDB Entry DOI: 10.7270/Q22R3VNT | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Genome polyprotein (Hepatitis C Virus) | BDBM50495935 (CHEMBL3120475) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 0.0900 | n/a | n/a | n/a | n/a | n/a | n/a |
Merck Research Laboratories Curated by ChEMBL | Assay Description Inhibition of Hepatitis C virus genotype 1b NS3/4A protease D168Y mutant assessed as substrate cleavage using Ac-C(Eu)DDMEEAbu(COO)ASK(QSY7)-amide as... | ACS Med Chem Lett 5: 264-9 (2014) Article DOI: 10.1021/ml400466p BindingDB Entry DOI: 10.7270/Q22R3VNT | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Genome polyprotein (Hepatitis C Virus) | BDBM50495941 (CHEMBL3349203) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 0.100 | n/a | n/a | n/a | n/a | n/a | n/a |
Merck Research Laboratories Curated by ChEMBL | Assay Description Inhibition of Hepatitis C virus genotype 1b NS3/4A protease R155K mutant assessed as substrate cleavage using Ac-C(Eu)DDMEEAbu(COO)ASK(QSY7)-amide as... | ACS Med Chem Lett 5: 264-9 (2014) Article DOI: 10.1021/ml400466p BindingDB Entry DOI: 10.7270/Q22R3VNT | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Genome polyprotein (Hepatitis C Virus) | BDBM50495942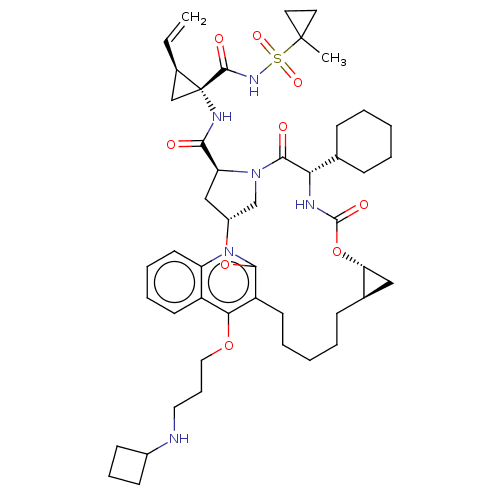 (CHEMBL3120488) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 0.100 | n/a | n/a | n/a | n/a | n/a | n/a |
Merck Research Laboratories Curated by ChEMBL | Assay Description Inhibition of Hepatitis C virus genotype 1b NS3/4A protease R155K mutant assessed as substrate cleavage using Ac-C(Eu)DDMEEAbu(COO)ASK(QSY7)-amide as... | ACS Med Chem Lett 5: 264-9 (2014) Article DOI: 10.1021/ml400466p BindingDB Entry DOI: 10.7270/Q22R3VNT | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Genome polyprotein (Hepatitis C Virus) | BDBM50495934 (CHEMBL3120476) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 0.110 | n/a | n/a | n/a | n/a | n/a | n/a |
Merck Research Laboratories Curated by ChEMBL | Assay Description Inhibition of Hepatitis C virus genotype 1b NS3/4A protease R155K mutant assessed as substrate cleavage using Ac-C(Eu)DDMEEAbu(COO)ASK(QSY7)-amide as... | ACS Med Chem Lett 5: 264-9 (2014) Article DOI: 10.1021/ml400466p BindingDB Entry DOI: 10.7270/Q22R3VNT | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Genome polyprotein (Hepatitis C Virus) | BDBM50495924 (CHEMBL3120477) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 0.110 | n/a | n/a | n/a | n/a | n/a | n/a |
Merck Research Laboratories Curated by ChEMBL | Assay Description Inhibition of Hepatitis C virus genotype 1b NS3/4A protease D168Y mutant assessed as substrate cleavage using Ac-C(Eu)DDMEEAbu(COO)ASK(QSY7)-amide as... | ACS Med Chem Lett 5: 264-9 (2014) Article DOI: 10.1021/ml400466p BindingDB Entry DOI: 10.7270/Q22R3VNT | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Genome polyprotein (Hepatitis C Virus) | BDBM50495932 (CHEMBL3349201) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 0.120 | n/a | n/a | n/a | n/a | n/a | n/a |
Merck Research Laboratories Curated by ChEMBL | Assay Description Inhibition of Hepatitis C virus genotype 1b NS3/4A protease R155K mutant assessed as substrate cleavage using Ac-C(Eu)DDMEEAbu(COO)ASK(QSY7)-amide as... | ACS Med Chem Lett 5: 264-9 (2014) Article DOI: 10.1021/ml400466p BindingDB Entry DOI: 10.7270/Q22R3VNT | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Genome polyprotein (Hepatitis C Virus) | BDBM50495928 (CHEMBL3349199) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 0.130 | n/a | n/a | n/a | n/a | n/a | n/a |
Merck Research Laboratories Curated by ChEMBL | Assay Description Inhibition of Hepatitis C virus genotype 1b NS3/4A protease A156V mutant assessed as substrate cleavage using Ac-C(Eu)DDMEEAbu(COO)ASK(QSY7)-amide as... | ACS Med Chem Lett 5: 264-9 (2014) Article DOI: 10.1021/ml400466p BindingDB Entry DOI: 10.7270/Q22R3VNT | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Genome polyprotein (Hepatitis C Virus) | BDBM50495926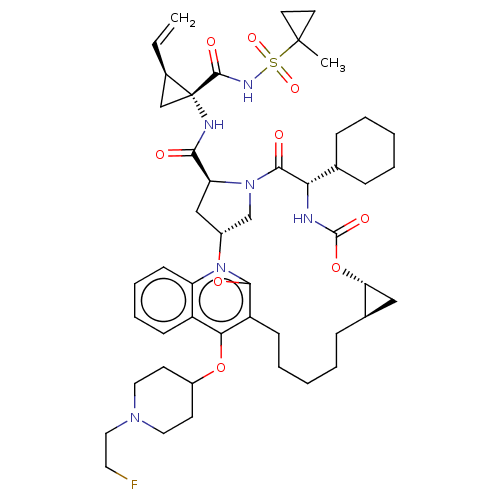 (CHEMBL3120482) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 0.130 | n/a | n/a | n/a | n/a | n/a | n/a |
Merck Research Laboratories Curated by ChEMBL | Assay Description Inhibition of Hepatitis C virus genotype 1b NS3/4A protease R155K mutant assessed as substrate cleavage using Ac-C(Eu)DDMEEAbu(COO)ASK(QSY7)-amide as... | ACS Med Chem Lett 5: 264-9 (2014) Article DOI: 10.1021/ml400466p BindingDB Entry DOI: 10.7270/Q22R3VNT | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Genome polyprotein (Hepatitis C Virus) | BDBM50485492 (Grazoprevir | Grazoprevir monohydrate | MK-5172 | ...) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | Purchase MCE PC cid PC sid PDB UniChem | PDB Article PubMed | n/a | n/a | 0.140 | n/a | n/a | n/a | n/a | n/a | n/a |
Merck Research Laboratories Curated by ChEMBL | Assay Description Inhibition of Hepatitis C virus genotype 1b NS3/4A protease D168Y mutant assessed as substrate cleavage using Ac-C(Eu)DDMEEAbu(COO)ASK(QSY7)-amide as... | ACS Med Chem Lett 5: 264-9 (2014) Article DOI: 10.1021/ml400466p BindingDB Entry DOI: 10.7270/Q22R3VNT | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Genome polyprotein (Hepatitis C Virus) | BDBM50495945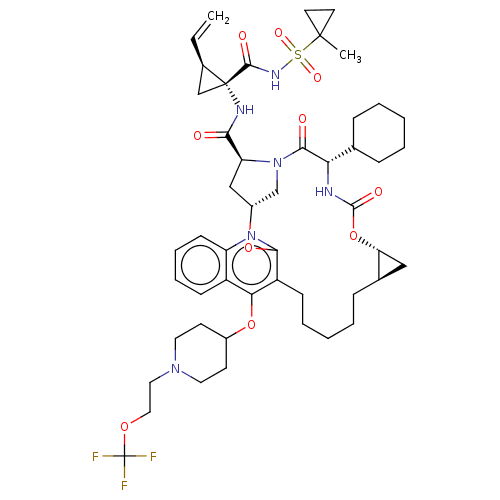 (CHEMBL3120479) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 0.150 | n/a | n/a | n/a | n/a | n/a | n/a |
Merck Research Laboratories Curated by ChEMBL | Assay Description Inhibition of Hepatitis C virus genotype 1b NS3/4A protease R155K mutant assessed as substrate cleavage using Ac-C(Eu)DDMEEAbu(COO)ASK(QSY7)-amide as... | ACS Med Chem Lett 5: 264-9 (2014) Article DOI: 10.1021/ml400466p BindingDB Entry DOI: 10.7270/Q22R3VNT | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Genome polyprotein (Hepatitis C Virus) | BDBM50495925 (CHEMBL3120478) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 0.150 | n/a | n/a | n/a | n/a | n/a | n/a |
Merck Research Laboratories Curated by ChEMBL | Assay Description Inhibition of Hepatitis C virus genotype 1b NS3/4A protease D168Y mutant assessed as substrate cleavage using Ac-C(Eu)DDMEEAbu(COO)ASK(QSY7)-amide as... | ACS Med Chem Lett 5: 264-9 (2014) Article DOI: 10.1021/ml400466p BindingDB Entry DOI: 10.7270/Q22R3VNT | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Genome polyprotein (Hepatitis C Virus) | BDBM50495938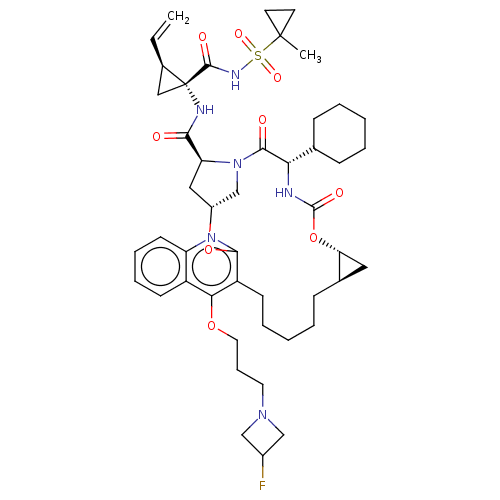 (CHEMBL3120489) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 0.160 | n/a | n/a | n/a | n/a | n/a | n/a |
Merck Research Laboratories Curated by ChEMBL | Assay Description Inhibition of Hepatitis C virus genotype 1b NS3/4A protease R155K mutant assessed as substrate cleavage using Ac-C(Eu)DDMEEAbu(COO)ASK(QSY7)-amide as... | ACS Med Chem Lett 5: 264-9 (2014) Article DOI: 10.1021/ml400466p BindingDB Entry DOI: 10.7270/Q22R3VNT | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Genome polyprotein (Hepatitis C Virus) | BDBM50495925 (CHEMBL3120478) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 0.180 | n/a | n/a | n/a | n/a | n/a | n/a |
Merck Research Laboratories Curated by ChEMBL | Assay Description Inhibition of Hepatitis C virus genotype 1b NS3/4A protease R155K mutant assessed as substrate cleavage using Ac-C(Eu)DDMEEAbu(COO)ASK(QSY7)-amide as... | ACS Med Chem Lett 5: 264-9 (2014) Article DOI: 10.1021/ml400466p BindingDB Entry DOI: 10.7270/Q22R3VNT | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Genome polyprotein (Hepatitis C Virus) | BDBM50495939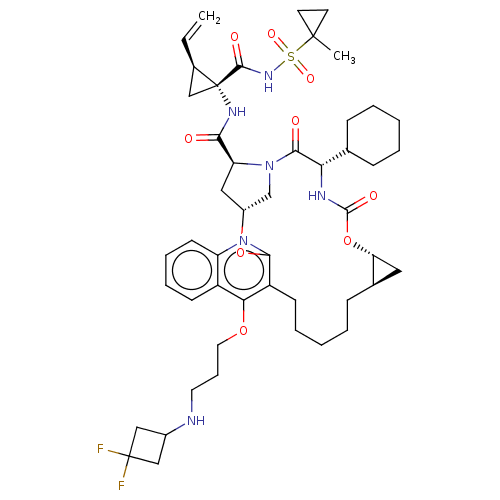 (CHEMBL3120169) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | n/a | n/a | 0.180 | n/a | n/a | n/a | n/a | n/a | n/a |
Merck Research Laboratories Curated by ChEMBL | Assay Description Inhibition of Hepatitis C virus genotype 1b NS3/4A protease R155K mutant assessed as substrate cleavage using Ac-C(Eu)DDMEEAbu(COO)ASK(QSY7)-amide as... | ACS Med Chem Lett 5: 264-9 (2014) Article DOI: 10.1021/ml400466p BindingDB Entry DOI: 10.7270/Q22R3VNT | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Displayed 1 to 50 (of 125 total ) | Next | Last >> |