

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Gag-Pol polyprotein [484-582,Q491K,I494L,I497V,V499I,E519D,R541K,D544E,Q553K,L573M] (Human immunodeficiency virus) | BDBM112660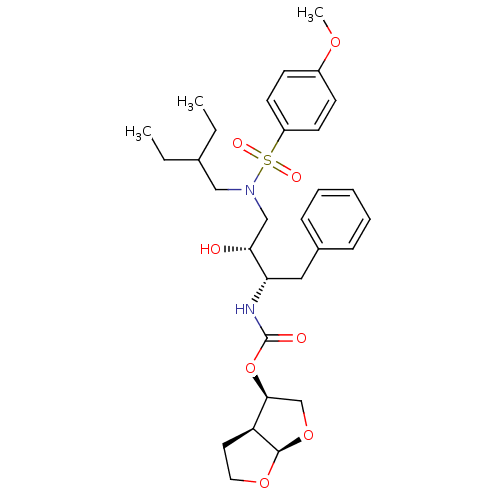 ((3R,3aS,6aR)-Hexahydrofuro[2,3-b]furan-3-yl ((2S,3...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid PDB UniChem Similars | PDB Article PubMed | 0.000200 | -72.5 | n/a | n/a | n/a | n/a | n/a | 4.7 | 25 |
University of Massachusetts Medical School, Worcester, MA 01605, USA | Assay Description The reaction mixture contained 2 µL of protease and 2 µL of inhibitor (or DMSO as a control) and was incubated for 20-30 min at room temper... | Chem Biol 20: 1116-24 (2013) Article DOI: 10.1016/j.chembiol.2013.07.014 BindingDB Entry DOI: 10.7270/Q2HQ3XKF | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Protease (Human immunodeficiency virus 1 (HIV-1)) | BDBM8125 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 0.000200 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Sanford-Burnham Medical Research Institute Curated by ChEMBL | Assay Description Inhibition of wild type HIV1 protease by FRET assay | J Med Chem 55: 6328-41 (2012) Article DOI: 10.1021/jm300238h BindingDB Entry DOI: 10.7270/Q28G8PKV | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Gag-Pol polyprotein [484-582,Q491K,I494L,I497V,V499I,E519D,R541K,D544E,Q553K,L573M] (Human immunodeficiency virus) | BDBM112656 ((3R,3aS,6aR)-Hexahydrofuro[2,3-b]furan-3-yl ((2S,3...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid PDB UniChem Similars | PDB Article PubMed | 0.000500 | -70.2 | n/a | n/a | n/a | n/a | n/a | 4.7 | 25 |
University of Massachusetts Medical School, Worcester, MA 01605, USA | Assay Description The reaction mixture contained 2 µL of protease and 2 µL of inhibitor (or DMSO as a control) and was incubated for 20-30 min at room temper... | Chem Biol 20: 1116-24 (2013) Article DOI: 10.1016/j.chembiol.2013.07.014 BindingDB Entry DOI: 10.7270/Q2HQ3XKF | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Gag-Pol polyprotein [484-582,Q491K,I494L,I497V,V499I,E519D,R541K,D544E,Q553K,L573M] (Human immunodeficiency virus) | BDBM112661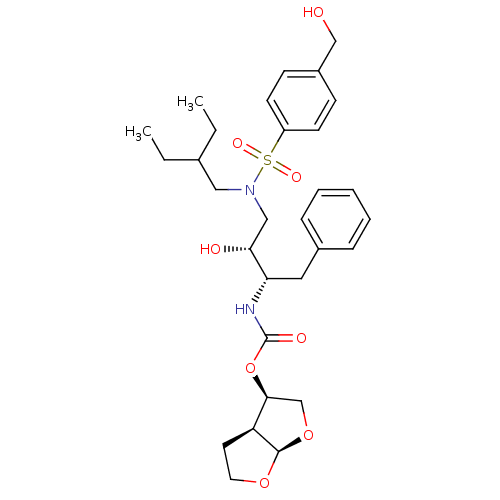 ((3R,3aS,6aR)-Hexahydrofuro[2,3-b]furan-3-yl ((2S,3...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid PDB UniChem Similars | PDB Article PubMed | 0.000500 | -70.2 | n/a | n/a | n/a | n/a | n/a | 4.7 | 25 |
University of Massachusetts Medical School, Worcester, MA 01605, USA | Assay Description The reaction mixture contained 2 µL of protease and 2 µL of inhibitor (or DMSO as a control) and was incubated for 20-30 min at room temper... | Chem Biol 20: 1116-24 (2013) Article DOI: 10.1016/j.chembiol.2013.07.014 BindingDB Entry DOI: 10.7270/Q2HQ3XKF | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| HIV-1 protease (Human immunodeficiency virus) | BDBM12877 ((5S)-3-(3-Acetylphenyl)-N-[(1S,2R)-1-benzyl-2-hydr...) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | 0.000794 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
UMBI Curated by ChEMBL | Assay Description Inhibition of HIV1 protease Q7K mutant by FRET method | J Med Chem 52: 737-54 (2009) Article DOI: 10.1021/jm8009525 BindingDB Entry DOI: 10.7270/Q2PN98FX | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Protease (Human immunodeficiency virus 1 (HIV-1)) | BDBM12877 ((5S)-3-(3-Acetylphenyl)-N-[(1S,2R)-1-benzyl-2-hydr...) | PDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | 0.000800 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
University of Massachusetts Medical School Curated by ChEMBL | Assay Description Inhibition of wild type HIV1 protease by FRET | J Med Chem 53: 7699-708 (2010) Article DOI: 10.1021/jm1008743 BindingDB Entry DOI: 10.7270/Q2BZ68WM | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Gag-Pol polyprotein [484-582,Q491K,I494L,I497V,V499I,E519D,R541K,D544E,Q553K,L573M] (Human immunodeficiency virus) | BDBM112662 ((3R,3aS,6aR)-Hexahydrofuro[2,3-b]furan-3-yl ((2S,3...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid PDB UniChem Similars | PDB Article PubMed | 0.000800 | -69.0 | n/a | n/a | n/a | n/a | n/a | 4.7 | 25 |
University of Massachusetts Medical School, Worcester, MA 01605, USA | Assay Description The reaction mixture contained 2 µL of protease and 2 µL of inhibitor (or DMSO as a control) and was incubated for 20-30 min at room temper... | Chem Biol 20: 1116-24 (2013) Article DOI: 10.1016/j.chembiol.2013.07.014 BindingDB Entry DOI: 10.7270/Q2HQ3XKF | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| HIV-1 protease (Human immunodeficiency virus) | BDBM12877 ((5S)-3-(3-Acetylphenyl)-N-[(1S,2R)-1-benzyl-2-hydr...) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | 0.000800 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
UMBI Curated by ChEMBL | Assay Description Inhibition of HIV1 protease Q7K mutant by FRET method | J Med Chem 52: 737-54 (2009) Article DOI: 10.1021/jm8009525 BindingDB Entry DOI: 10.7270/Q2PN98FX | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Dimer of Gag-Pol polyprotein [491-589,Q496K] (Human immunodeficiency virus type 1) | BDBM12877 ((5S)-3-(3-Acetylphenyl)-N-[(1S,2R)-1-benzyl-2-hydr...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | 0.000800 | -70.2 | n/a | n/a | n/a | n/a | n/a | 4.7 | 30 |
University of Massachusetts Medical School | Assay Description HIV-1 protease inhibitor activities were determined by the fluorescence resonance energy transfer (FRET) method. The energy transfer donor (EDANS) an... | J Med Chem 49: 7342-56 (2006) Article DOI: 10.1021/jm060666p BindingDB Entry DOI: 10.7270/Q25T3HQ6 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Gag-Pol polyprotein [484-582,Q491K,I494L,I497V,V499I,E519D,R541K,D544E,Q553K,L573M] (Human immunodeficiency virus) | BDBM112657 ((3R,3aS,6aR)-Hexahydrofuro[2,3-b]furan-3-yl ((2S,3...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid PDB UniChem Similars | PDB Article PubMed | 0.000900 | -68.8 | n/a | n/a | n/a | n/a | n/a | 4.7 | 25 |
University of Massachusetts Medical School, Worcester, MA 01605, USA | Assay Description The reaction mixture contained 2 µL of protease and 2 µL of inhibitor (or DMSO as a control) and was incubated for 20-30 min at room temper... | Chem Biol 20: 1116-24 (2013) Article DOI: 10.1016/j.chembiol.2013.07.014 BindingDB Entry DOI: 10.7270/Q2HQ3XKF | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Protease (Human immunodeficiency virus 1 (HIV-1)) | BDBM12877 ((5S)-3-(3-Acetylphenyl)-N-[(1S,2R)-1-benzyl-2-hydr...) | PDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | 0.00100 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Sanford-Burnham Medical Research Institute Curated by ChEMBL | Assay Description Inhibition of wild type HIV1 protease by FRET assay | J Med Chem 55: 6328-41 (2012) Article DOI: 10.1021/jm300238h BindingDB Entry DOI: 10.7270/Q28G8PKV | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| HIV-1 protease (Human immunodeficiency virus) | BDBM8125 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 0.00100 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Sanford-Burnham Medical Research Institute Curated by ChEMBL | Assay Description Inhibition of HIV1 protease I50V, A71V mutant by FRET assay | J Med Chem 55: 6328-41 (2012) Article DOI: 10.1021/jm300238h BindingDB Entry DOI: 10.7270/Q28G8PKV | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Induced myeloid leukemia cell differentiation protein Mcl-1 (Homo sapiens (Human)) | BDBM451126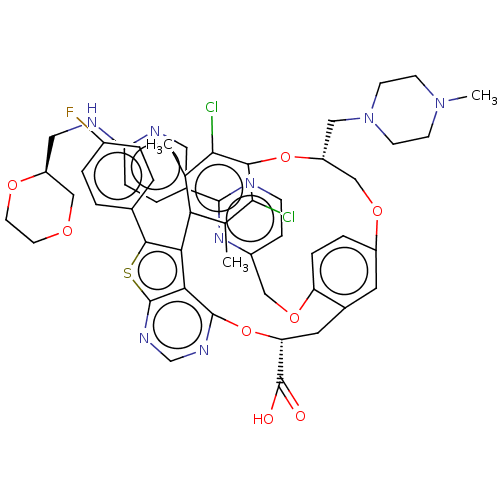 (US10676485, Example 36) | PDB MMDB NCI pathway KEGG UniProtKB/SwissProt B.MOAD antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem | US Patent | 0.00100 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
AbbVie Inc. US Patent | Assay Description The ability of the exemplary MCL-1 inhibitors of Examples 1 through 151 to bind MCL-1 was demonstrated using the Time Resolved-Fluorescence Resonance... | US Patent US10676485 (2020) BindingDB Entry DOI: 10.7270/Q2PZ5CWK | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Protease (Human immunodeficiency virus 1 (HIV-1)) | BDBM50485746 (CHEMBL2165886) | PDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 0.00100 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Sanford-Burnham Medical Research Institute Curated by ChEMBL | Assay Description Inhibition of wild type HIV1 protease by FRET assay | J Med Chem 55: 6328-41 (2012) Article DOI: 10.1021/jm300238h BindingDB Entry DOI: 10.7270/Q28G8PKV | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dihydrofolate reductase (Escherichia coli) | BDBM18050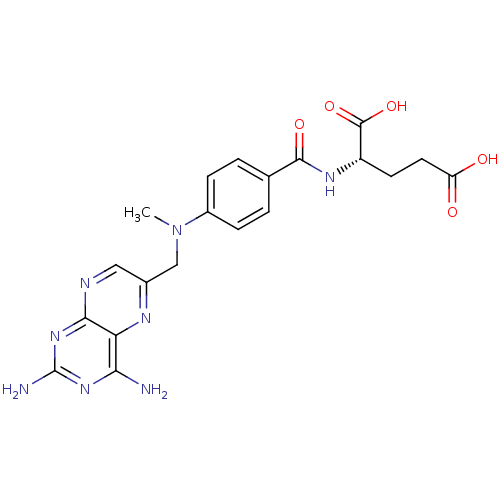 (2-[(4-{[(2,4-diaminopteridin-6-yl)methyl](methyl)a...) | MMDB NCI pathway Reactome pathway KEGG UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE KEGG MMDB PC cid PC sid PDB UniChem Patents Similars | Article PubMed | 0.00100 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
University of Connecticut Curated by ChEMBL | Assay Description Inhibition of recombinant Escherichia coli DHFR | ACS Med Chem Lett 7: 692-6 (2016) Article DOI: 10.1021/acsmedchemlett.6b00120 BindingDB Entry DOI: 10.7270/Q2DJ5HKK | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Gag-Pol polyprotein [484-582,Q491K,I497V,V499I,E519D,G532V,I538V,R541K,D544E,L547P,Q553K,V566A,L573M] (Human immunodeficiency virus) | BDBM112660 ((3R,3aS,6aR)-Hexahydrofuro[2,3-b]furan-3-yl ((2S,3...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid PDB UniChem Similars | Article PubMed | 0.00100 | -68.5 | n/a | n/a | n/a | n/a | n/a | 4.7 | 25 |
University of Massachusetts Medical School, Worcester, MA 01605, USA | Assay Description The reaction mixture contained 2 µL of protease and 2 µL of inhibitor (or DMSO as a control) and was incubated for 20-30 min at room temper... | Chem Biol 20: 1116-24 (2013) Article DOI: 10.1016/j.chembiol.2013.07.014 BindingDB Entry DOI: 10.7270/Q2HQ3XKF | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Gag-Pol polyprotein [484-582,Q491K,I494L,I497V,V499I,E519D,R541K,D544E,Q553K,L573M] (Human immunodeficiency virus) | BDBM112663 ((3R,3aS,6aR)-Hexahydrofuro[2,3-b]furan-3-yl ((2S,3...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid PDB UniChem Similars | PDB Article PubMed | 0.00150 | -67.5 | n/a | n/a | n/a | n/a | n/a | 4.7 | 25 |
University of Massachusetts Medical School, Worcester, MA 01605, USA | Assay Description The reaction mixture contained 2 µL of protease and 2 µL of inhibitor (or DMSO as a control) and was incubated for 20-30 min at room temper... | Chem Biol 20: 1116-24 (2013) Article DOI: 10.1016/j.chembiol.2013.07.014 BindingDB Entry DOI: 10.7270/Q2HQ3XKF | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Dihydrofolate reductase (Homo sapiens (Human)) | BDBM18050 (2-[(4-{[(2,4-diaminopteridin-6-yl)methyl](methyl)a...) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE KEGG MMDB PC cid PC sid PDB UniChem Patents Similars | DrugBank MMDB PDB Article PubMed | 0.00300 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
University of Connecticut Curated by ChEMBL | Assay Description Inhibition of recombinant human DHFR assessed as reduction in NADPH oxidation using dihydrofolate as substrate by fluorescence spectrophotometric ana... | ACS Med Chem Lett 7: 692-6 (2016) Article DOI: 10.1021/acsmedchemlett.6b00120 BindingDB Entry DOI: 10.7270/Q2DJ5HKK | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Induced myeloid leukemia cell differentiation protein Mcl-1 (Homo sapiens (Human)) | BDBM451115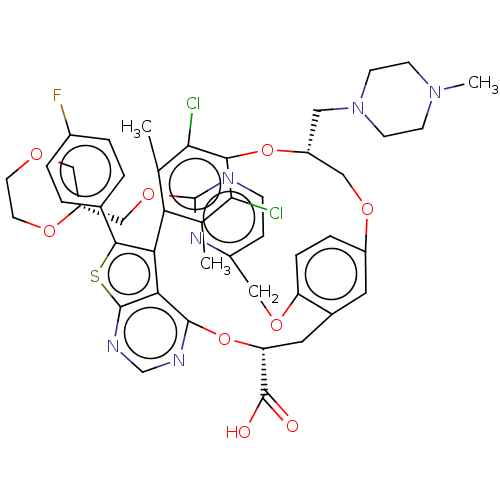 (US10676485, Example 25) | PDB MMDB NCI pathway KEGG UniProtKB/SwissProt B.MOAD antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem | US Patent | 0.00300 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
AbbVie Inc. US Patent | Assay Description The ability of the exemplary MCL-1 inhibitors of Examples 1 through 151 to bind MCL-1 was demonstrated using the Time Resolved-Fluorescence Resonance... | US Patent US10676485 (2020) BindingDB Entry DOI: 10.7270/Q2PZ5CWK | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Protease (Human immunodeficiency virus 1 (HIV-1)) | BDBM50483092 (CHEMBL1276087) | PDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 0.00300 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
University of Massachusetts Medical School Curated by ChEMBL | Assay Description Inhibition of wild type HIV1 protease by FRET | J Med Chem 53: 7699-708 (2010) Article DOI: 10.1021/jm1008743 BindingDB Entry DOI: 10.7270/Q2BZ68WM | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Protease (Human immunodeficiency virus 1 (HIV-1)) | BDBM50485764 (CHEMBL2165912) | PDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid PDB UniChem | Article PubMed | 0.00300 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Sanford-Burnham Medical Research Institute Curated by ChEMBL | Assay Description Inhibition of wild type HIV1 protease by FRET assay | J Med Chem 55: 6328-41 (2012) Article DOI: 10.1021/jm300238h BindingDB Entry DOI: 10.7270/Q28G8PKV | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Protease (Human immunodeficiency virus 1 (HIV-1)) | BDBM50485744 (CHEMBL2165917) | PDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 0.00300 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Sanford-Burnham Medical Research Institute Curated by ChEMBL | Assay Description Inhibition of wild type HIV1 protease by FRET assay | J Med Chem 55: 6328-41 (2012) Article DOI: 10.1021/jm300238h BindingDB Entry DOI: 10.7270/Q28G8PKV | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Protease (Human immunodeficiency virus 1 (HIV-1)) | BDBM50485740 (CHEMBL2165903) | PDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid PDB UniChem | Article PubMed | 0.00300 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Sanford-Burnham Medical Research Institute Curated by ChEMBL | Assay Description Inhibition of wild type HIV1 protease by FRET assay | J Med Chem 55: 6328-41 (2012) Article DOI: 10.1021/jm300238h BindingDB Entry DOI: 10.7270/Q28G8PKV | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Adenosine receptor A2a (Homo sapiens (Human)) | BDBM50550812 (CHEMBL4756283) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 0.00380 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA | Assay Description Displacement of [3H]MSX2 from human adenosine receptor A2A expressed in HEK293 cell membranes incubated for 30 mins by radioligand competition assay | Citation and Details Article DOI: 10.1016/j.ejmech.2019.111879 BindingDB Entry DOI: 10.7270/Q24T6P0P | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| HIV-1 protease (Human immunodeficiency virus) | BDBM12878 ((5S)-3-(4-Acetylphenyl)-N-[(1S,2R)-1-benzyl-2-hydr...) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | 0.00398 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
UMBI Curated by ChEMBL | Assay Description Inhibition of HIV1 protease Q7K mutant by FRET method | J Med Chem 52: 737-54 (2009) Article DOI: 10.1021/jm8009525 BindingDB Entry DOI: 10.7270/Q2PN98FX | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| HIV-1 protease (Human immunodeficiency virus) | BDBM8125 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 0.00400 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Sanford-Burnham Medical Research Institute Curated by ChEMBL | Assay Description Inhibition of HIV1 protease L10I, G48V, I54V, L63P and V82A mutant by FRET assay | J Med Chem 55: 6328-41 (2012) Article DOI: 10.1021/jm300238h BindingDB Entry DOI: 10.7270/Q28G8PKV | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Gag-Pol polyprotein [484-582,Q491K,I497V,V499I,E519D,R541K,D544E,L547P,Q553K,A555V,G557S,I568V,L573M,L574M] (Human immunodeficiency virus) | BDBM112656 ((3R,3aS,6aR)-Hexahydrofuro[2,3-b]furan-3-yl ((2S,3...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid PDB UniChem Similars | PDB Article PubMed | 0.00400 | -65.1 | n/a | n/a | n/a | n/a | n/a | 4.7 | 25 |
University of Massachusetts Medical School, Worcester, MA 01605, USA | Assay Description The reaction mixture contained 2 µL of protease and 2 µL of inhibitor (or DMSO as a control) and was incubated for 20-30 min at room temper... | Chem Biol 20: 1116-24 (2013) Article DOI: 10.1016/j.chembiol.2013.07.014 BindingDB Entry DOI: 10.7270/Q2HQ3XKF | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Induced myeloid leukemia cell differentiation protein Mcl-1 (Homo sapiens (Human)) | BDBM451129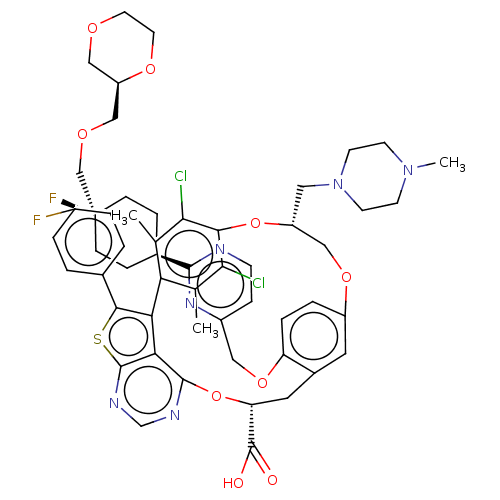 (US10676485, Example 39) | PDB MMDB NCI pathway KEGG UniProtKB/SwissProt B.MOAD antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem | US Patent | 0.00400 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
AbbVie Inc. US Patent | Assay Description The ability of the exemplary MCL-1 inhibitors of Examples 1 through 151 to bind MCL-1 was demonstrated using the Time Resolved-Fluorescence Resonance... | US Patent US10676485 (2020) BindingDB Entry DOI: 10.7270/Q2PZ5CWK | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [491-589,Q496K] (Human immunodeficiency virus type 1) | BDBM12878 ((5S)-3-(4-Acetylphenyl)-N-[(1S,2R)-1-benzyl-2-hydr...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | 0.00400 | -66.1 | n/a | n/a | n/a | n/a | n/a | 4.7 | 30 |
University of Massachusetts Medical School | Assay Description HIV-1 protease inhibitor activities were determined by the fluorescence resonance energy transfer (FRET) method. The energy transfer donor (EDANS) an... | J Med Chem 49: 7342-56 (2006) Article DOI: 10.1021/jm060666p BindingDB Entry DOI: 10.7270/Q25T3HQ6 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| HIV-1 protease (Human immunodeficiency virus) | BDBM12878 ((5S)-3-(4-Acetylphenyl)-N-[(1S,2R)-1-benzyl-2-hydr...) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | MMDB PC cid PC sid PDB UniChem Similars | PDB Article PubMed | 0.00400 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
UMBI Curated by ChEMBL | Assay Description Inhibition of HIV1 protease Q7K mutant by FRET method | J Med Chem 52: 737-54 (2009) Article DOI: 10.1021/jm8009525 BindingDB Entry DOI: 10.7270/Q2PN98FX | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Adenosine receptor A2a (Homo sapiens (Human)) | BDBM50493145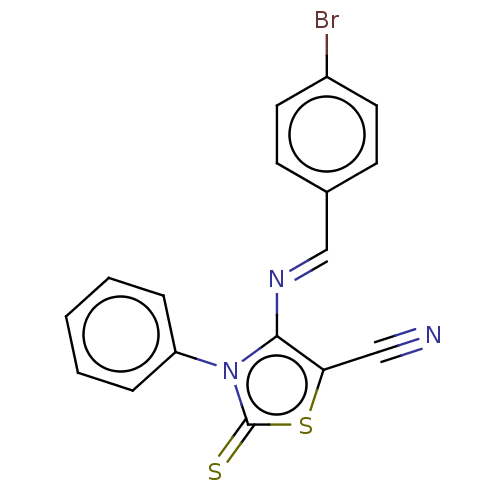 (CHEMBL2419149) | PDB MMDB NCI pathway Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 0.00420 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA | Assay Description Displacement of [3H]MSX2 from human adenosine receptor A2A expressed in HEK293 cell membranes incubated for 30 mins by radioligand competition assay | Citation and Details Article DOI: 10.1016/j.ejmech.2019.111879 BindingDB Entry DOI: 10.7270/Q24T6P0P | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Protease (Human immunodeficiency virus 1 (HIV-1)) | BDBM50485760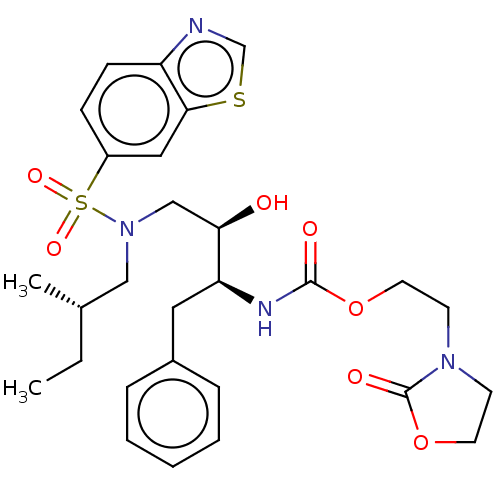 (CHEMBL2165901) | PDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 0.00500 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Sanford-Burnham Medical Research Institute Curated by ChEMBL | Assay Description Inhibition of wild type HIV1 protease by FRET assay | J Med Chem 55: 6328-41 (2012) Article DOI: 10.1021/jm300238h BindingDB Entry DOI: 10.7270/Q28G8PKV | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Protease (Human immunodeficiency virus 1 (HIV-1)) | BDBM50180655 (A-157378-0 | A-157378.0 | ABT-378 | CHEBI:31781 | ...) | PDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | Purchase DrugBank MCE KEGG PC cid PC sid PDB UniChem Similars | PDB Article PubMed | 0.00500 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
University of Massachusetts Medical School Curated by ChEMBL | Assay Description Inhibition of wild type HIV1 protease by FRET | J Med Chem 53: 7699-708 (2010) Article DOI: 10.1021/jm1008743 BindingDB Entry DOI: 10.7270/Q2BZ68WM | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Protease (Human immunodeficiency virus 1 (HIV-1)) | BDBM112661 ((3R,3aS,6aR)-Hexahydrofuro[2,3-b]furan-3-yl ((2S,3...) | PDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid PDB UniChem Similars | Article PubMed | 0.00500 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
University of Massachusetts Medical School Curated by ChEMBL | Assay Description Inhibition of HIV1 NL4-3 protease I84V mutant expressed in Escherichia coli TAP-106 cells using EDANS/DABCYL-labelled 10-amino acid containing natura... | J Med Chem 62: 8062-8079 (2019) Article DOI: 10.1021/acs.jmedchem.9b00838 BindingDB Entry DOI: 10.7270/Q26H4MQ1 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Induced myeloid leukemia cell differentiation protein Mcl-1 (Homo sapiens (Human)) | BDBM451099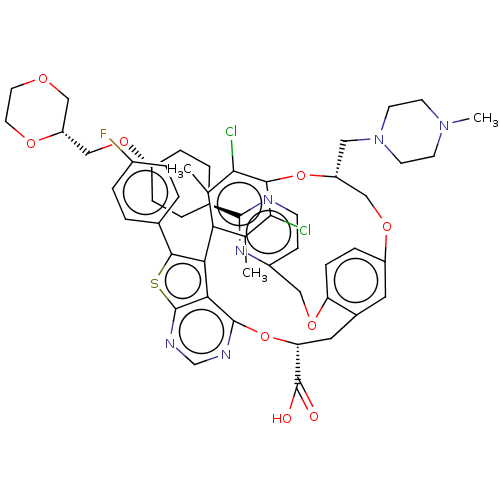 (US10676485, Example 9) | PDB MMDB NCI pathway KEGG UniProtKB/SwissProt B.MOAD antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem | US Patent | 0.00500 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
AbbVie Inc. US Patent | Assay Description The ability of the exemplary MCL-1 inhibitors of Examples 1 through 151 to bind MCL-1 was demonstrated using the Time Resolved-Fluorescence Resonance... | US Patent US10676485 (2020) BindingDB Entry DOI: 10.7270/Q2PZ5CWK | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Gag-Pol polyprotein [489-587] (Human immunodeficiency virus type 1) | BDBM8125 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE MMDB PC cid PC sid PDB UniChem Patents Similars | DrugBank PDB Article PubMed | <0.00500 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
University of Massachusetts Medical School Curated by ChEMBL | Assay Description Inhibition of HIV1 NL4-3 protease expressed in Escherichia coli TAP-106 cells using EDANS/DABCYL-labelled 10-amino acid containing protease cleavage ... | J Med Chem 63: 8296-8313 (2020) Article DOI: 10.1021/acs.jmedchem.0c00529 BindingDB Entry DOI: 10.7270/Q2959N46 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Gag-Pol polyprotein [489-587] (Human immunodeficiency virus type 1) | BDBM50180655 (A-157378-0 | A-157378.0 | ABT-378 | CHEBI:31781 | ...) | PDB MMDB UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase DrugBank MCE KEGG PC cid PC sid PDB UniChem Similars | DrugBank PDB Article PubMed | <0.00500 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
University of Massachusetts Medical School Curated by ChEMBL | Assay Description Inhibition of HIV1 NL4-3 protease expressed in Escherichia coli TAP-106 cells using EDANS/DABCYL-labelled 10-amino acid containing protease cleavage ... | J Med Chem 63: 8296-8313 (2020) Article DOI: 10.1021/acs.jmedchem.0c00529 BindingDB Entry DOI: 10.7270/Q2959N46 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Protease (Human immunodeficiency virus 1 (HIV-1)) | BDBM50485747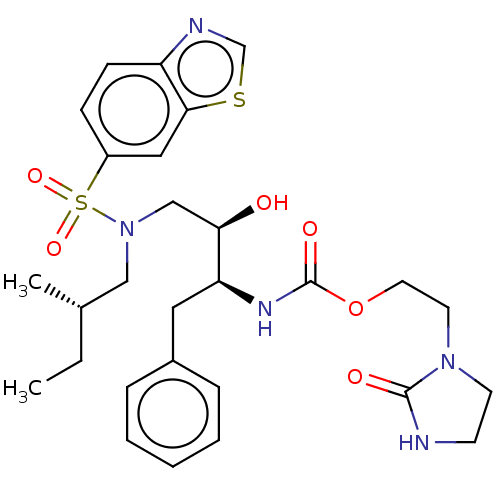 (CHEMBL2165900) | PDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 0.00500 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Sanford-Burnham Medical Research Institute Curated by ChEMBL | Assay Description Inhibition of wild type HIV1 protease by FRET assay | J Med Chem 55: 6328-41 (2012) Article DOI: 10.1021/jm300238h BindingDB Entry DOI: 10.7270/Q28G8PKV | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Gag-Pol polyprotein [484-582,Q491K,I497V,V499I,E519D,G532V,I538V,R541K,D544E,L547P,Q553K,V566A,L573M] (Human immunodeficiency virus) | BDBM112661 ((3R,3aS,6aR)-Hexahydrofuro[2,3-b]furan-3-yl ((2S,3...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid PDB UniChem Similars | Article PubMed | 0.00500 | -64.5 | n/a | n/a | n/a | n/a | n/a | 4.7 | 25 |
University of Massachusetts Medical School, Worcester, MA 01605, USA | Assay Description The reaction mixture contained 2 µL of protease and 2 µL of inhibitor (or DMSO as a control) and was incubated for 20-30 min at room temper... | Chem Biol 20: 1116-24 (2013) Article DOI: 10.1016/j.chembiol.2013.07.014 BindingDB Entry DOI: 10.7270/Q2HQ3XKF | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Gag-Pol polyprotein [484-582,Q491K,I497V,V499I,E519D,G532V,I538V,R541K,D544E,L547P,Q553K,V566A,L573M] (Human immunodeficiency virus) | BDBM112655 ((3R,3aS,6aR)-Hexahydrofuro[2,3-b]furan-3-yl ((2S,3...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid PDB UniChem Similars | Article PubMed | 0.00500 | -64.5 | n/a | n/a | n/a | n/a | n/a | 4.7 | 25 |
University of Massachusetts Medical School, Worcester, MA 01605, USA | Assay Description The reaction mixture contained 2 µL of protease and 2 µL of inhibitor (or DMSO as a control) and was incubated for 20-30 min at room temper... | Chem Biol 20: 1116-24 (2013) Article DOI: 10.1016/j.chembiol.2013.07.014 BindingDB Entry DOI: 10.7270/Q2HQ3XKF | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [491-589,Q496K] (Human immunodeficiency virus type 1) | BDBM578 ((2S)-N-[(2S,4S,5S)-5-[2-(2,6-dimethylphenoxy)aceta...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL MCE MMDB PC cid PC sid PDB UniChem Patents Similars | MMDB PDB Article PubMed | 0.00500 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
University of Massachusetts Medical School | Assay Description HIV-1 protease inhibitor activities were determined by the fluorescence resonance energy transfer (FRET) method. The energy transfer donor (EDANS) an... | J Med Chem 49: 7342-56 (2006) Article DOI: 10.1021/jm060666p BindingDB Entry DOI: 10.7270/Q25T3HQ6 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| HIV-1 protease (Human immunodeficiency virus) | BDBM578 ((2S)-N-[(2S,4S,5S)-5-[2-(2,6-dimethylphenoxy)aceta...) | PDB MMDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | CHEMBL MCE MMDB PC cid PC sid PDB UniChem Patents Similars | MMDB PDB Article PubMed | 0.00500 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
University of Maryland | Assay Description Inhibition assay using HIV protease and Sulfonamide compounds. | Chem Biol Drug Des 69: 298-313 (2007) Article DOI: 10.1111/j.1747-0285.2007.00514.x BindingDB Entry DOI: 10.7270/Q2TQ6011 | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Gag-Pol polyprotein [484-582,Q491K,I494L,I497V,V499I,E519D,R541K,D544E,Q553K,L573M] (Human immunodeficiency virus) | BDBM8125 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 0.00500 | -64.5 | n/a | n/a | n/a | n/a | n/a | 4.7 | 25 |
University of Massachusetts Medical School, Worcester, MA 01605, USA | Assay Description The reaction mixture contained 2 µL of protease and 2 µL of inhibitor (or DMSO as a control) and was incubated for 20-30 min at room temper... | Chem Biol 20: 1116-24 (2013) Article DOI: 10.1016/j.chembiol.2013.07.014 BindingDB Entry DOI: 10.7270/Q2HQ3XKF | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Gag-Pol polyprotein [484-582,Q491K,I494L,I497V,V499I,E519D,R541K,D544E,Q553K,L573M] (Human immunodeficiency virus) | BDBM112659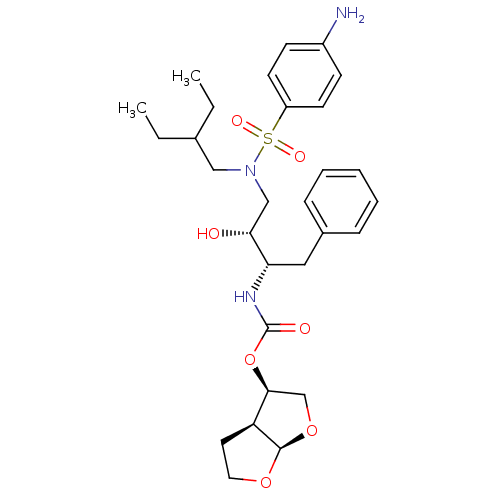 ((3R,3aS,6aR)-Hexahydrofuro[2,3-b]furan-3-yl ((2S,3...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid PDB UniChem Similars | PDB Article PubMed | 0.00500 | -64.5 | n/a | n/a | n/a | n/a | n/a | 4.7 | 25 |
University of Massachusetts Medical School, Worcester, MA 01605, USA | Assay Description The reaction mixture contained 2 µL of protease and 2 µL of inhibitor (or DMSO as a control) and was incubated for 20-30 min at room temper... | Chem Biol 20: 1116-24 (2013) Article DOI: 10.1016/j.chembiol.2013.07.014 BindingDB Entry DOI: 10.7270/Q2HQ3XKF | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Gag-Pol polyprotein [484-582,Q491K,I494L,I497V,V499I,E519D,I534V,R541K,D544E,Q553K,A555V,L573M] (Human immunodeficiency virus) | BDBM112662 ((3R,3aS,6aR)-Hexahydrofuro[2,3-b]furan-3-yl ((2S,3...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid PDB UniChem Similars | PDB Article PubMed | 0.00600 | -64.1 | n/a | n/a | n/a | n/a | n/a | 4.7 | 25 |
University of Massachusetts Medical School, Worcester, MA 01605, USA | Assay Description The reaction mixture contained 2 µL of protease and 2 µL of inhibitor (or DMSO as a control) and was incubated for 20-30 min at room temper... | Chem Biol 20: 1116-24 (2013) Article DOI: 10.1016/j.chembiol.2013.07.014 BindingDB Entry DOI: 10.7270/Q2HQ3XKF | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Dimer of Gag-Pol polyprotein [491-589,Q496K] (Human immunodeficiency virus type 1) | BDBM12883 ((5S)-3-(3-Acetylphenyl)-N-[(1S,2R)-3-[[(benzo[1,3]...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | MMDB PC cid PC sid PDB UniChem Patents Similars | MMDB Article PubMed | 0.00600 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
University of Massachusetts Medical School | Assay Description HIV-1 protease inhibitor activities were determined by the fluorescence resonance energy transfer (FRET) method. The energy transfer donor (EDANS) an... | J Med Chem 49: 7342-56 (2006) Article DOI: 10.1021/jm060666p BindingDB Entry DOI: 10.7270/Q25T3HQ6 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| HIV-1 protease (Human immunodeficiency virus) | BDBM12883 ((5S)-3-(3-Acetylphenyl)-N-[(1S,2R)-3-[[(benzo[1,3]...) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | MMDB PC cid PC sid PDB UniChem Patents Similars | Article PubMed | 0.00600 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
UMBI Curated by ChEMBL | Assay Description Inhibition of HIV1 protease Q7K mutant by FRET method | J Med Chem 52: 737-54 (2009) Article DOI: 10.1021/jm8009525 BindingDB Entry DOI: 10.7270/Q2PN98FX | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| HIV-1 protease (Human immunodeficiency virus) | BDBM12885 ((5S)-N-[(1S,2R)-1-Benzyl-2-hydroxy-3-[isobutyl[(4-...) | PDB UniProtKB/TrEMBL GoogleScholar AffyNet | MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | 0.00600 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
UMBI Curated by ChEMBL | Assay Description Inhibition of HIV1 protease Q7K mutant by FRET method | J Med Chem 52: 737-54 (2009) Article DOI: 10.1021/jm8009525 BindingDB Entry DOI: 10.7270/Q2PN98FX | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Gag-Pol polyprotein [484-582,Q491K,I494L,I497V,V499I,E519D,I534V,R541K,D544E,Q553K,A555V,L573M] (Human immunodeficiency virus) | BDBM112654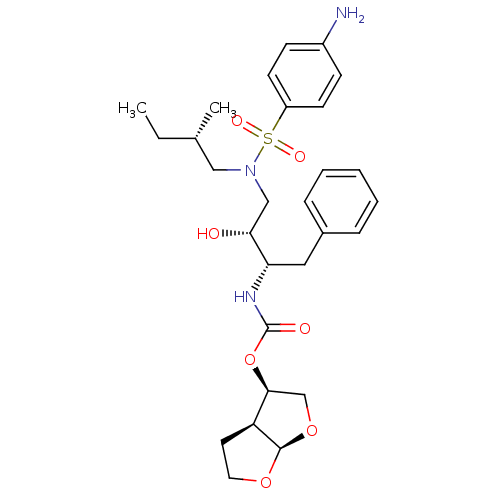 ((3R,3aS,6aR)-Hexahydrofuro[2,3-b]furan-3-yl ((2S,3...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid PDB UniChem Similars | PDB Article PubMed | 0.00600 | -64.1 | n/a | n/a | n/a | n/a | n/a | 4.7 | 25 |
University of Massachusetts Medical School, Worcester, MA 01605, USA | Assay Description The reaction mixture contained 2 µL of protease and 2 µL of inhibitor (or DMSO as a control) and was incubated for 20-30 min at room temper... | Chem Biol 20: 1116-24 (2013) Article DOI: 10.1016/j.chembiol.2013.07.014 BindingDB Entry DOI: 10.7270/Q2HQ3XKF | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Dimer of Gag-Pol polyprotein [491-589,Q496K] (Human immunodeficiency virus type 1) | BDBM12885 ((5S)-N-[(1S,2R)-1-Benzyl-2-hydroxy-3-[isobutyl[(4-...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | MMDB PC cid PC sid PDB UniChem Similars | MMDB Article PubMed | 0.00600 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
University of Massachusetts Medical School | Assay Description HIV-1 protease inhibitor activities were determined by the fluorescence resonance energy transfer (FRET) method. The energy transfer donor (EDANS) an... | J Med Chem 49: 7342-56 (2006) Article DOI: 10.1021/jm060666p BindingDB Entry DOI: 10.7270/Q25T3HQ6 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Displayed 1 to 50 (of 41369 total ) | Next | Last >> |