| Citation |  Deaton, DN; Do, Y; Holt, J; Jeune, MR; Kramer, HF; Larkin, AL; Orband-Miller, LA; Peckham, GE; Poole, C; Price, DJ; Schaller, LT; Shen, Y; Shewchuk, LM; Stewart, EL; Stuart, JD; Thomson, SA; Ward, P; Wilson, JW; Xu, T; Guss, JH; Musetti, C; Rendina, AR; Affleck, K; Anders, D; Hancock, AP; Hobbs, H; Hodgson, ST; Hutchinson, J; Leveridge, MV; Nicholls, H; Smith, IED; Somers, DO; Sneddon, HF; Uddin, S; Cleasby, A; Mortenson, PN; Richardson, C; Saxty, G The discovery of quinoline-3-carboxamides as hematopoietic prostaglandin D synthase (H-PGDS) inhibitors. Bioorg Med Chem27:1456-1478 (2019) [PubMed] Article Deaton, DN; Do, Y; Holt, J; Jeune, MR; Kramer, HF; Larkin, AL; Orband-Miller, LA; Peckham, GE; Poole, C; Price, DJ; Schaller, LT; Shen, Y; Shewchuk, LM; Stewart, EL; Stuart, JD; Thomson, SA; Ward, P; Wilson, JW; Xu, T; Guss, JH; Musetti, C; Rendina, AR; Affleck, K; Anders, D; Hancock, AP; Hobbs, H; Hodgson, ST; Hutchinson, J; Leveridge, MV; Nicholls, H; Smith, IED; Somers, DO; Sneddon, HF; Uddin, S; Cleasby, A; Mortenson, PN; Richardson, C; Saxty, G The discovery of quinoline-3-carboxamides as hematopoietic prostaglandin D synthase (H-PGDS) inhibitors. Bioorg Med Chem27:1456-1478 (2019) [PubMed] Article |
|---|
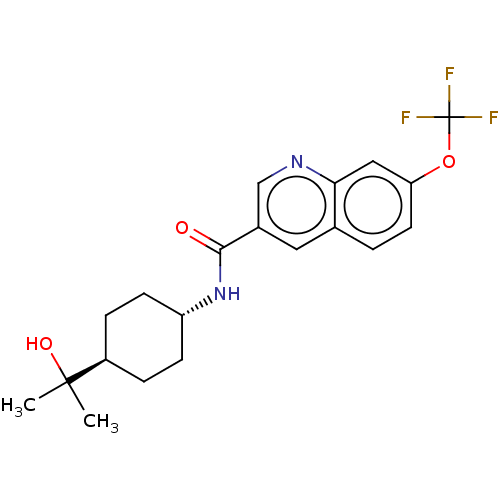
| SMILES | CC(C)(O)[C@H]1CC[C@@H](CC1)NC(=O)c1cnc2cc(OC(F)(F)F)ccc2c1 |r,wU:7.10,wD:4.3,(60.01,-18.28,;60.78,-19.61,;61.55,-18.27,;62.13,-20.38,;59.46,-20.39,;58.12,-19.63,;56.79,-20.41,;56.81,-21.94,;58.14,-22.71,;59.47,-21.93,;55.48,-22.72,;54.14,-21.96,;54.13,-20.42,;52.82,-22.74,;52.83,-24.29,;51.49,-25.06,;50.16,-24.3,;48.83,-25.08,;47.49,-24.31,;46.16,-25.07,;44.83,-24.3,;43.61,-23.6,;44.57,-22.72,;43.32,-24.88,;47.5,-22.76,;48.82,-21.99,;50.16,-22.75,;51.48,-21.98,)| |
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Deaton, DN; Do, Y; Holt, J; Jeune, MR; Kramer, HF; Larkin, AL; Orband-Miller, LA; Peckham, GE; Poole, C; Price, DJ; Schaller, LT; Shen, Y; Shewchuk, LM; Stewart, EL; Stuart, JD; Thomson, SA; Ward, P; Wilson, JW; Xu, T; Guss, JH; Musetti, C; Rendina, AR; Affleck, K; Anders, D; Hancock, AP; Hobbs, H; Hodgson, ST; Hutchinson, J; Leveridge, MV; Nicholls, H; Smith, IED; Somers, DO; Sneddon, HF; Uddin, S; Cleasby, A; Mortenson, PN; Richardson, C; Saxty, G The discovery of quinoline-3-carboxamides as hematopoietic prostaglandin D synthase (H-PGDS) inhibitors. Bioorg Med Chem27:1456-1478 (2019) [PubMed] Article
Deaton, DN; Do, Y; Holt, J; Jeune, MR; Kramer, HF; Larkin, AL; Orband-Miller, LA; Peckham, GE; Poole, C; Price, DJ; Schaller, LT; Shen, Y; Shewchuk, LM; Stewart, EL; Stuart, JD; Thomson, SA; Ward, P; Wilson, JW; Xu, T; Guss, JH; Musetti, C; Rendina, AR; Affleck, K; Anders, D; Hancock, AP; Hobbs, H; Hodgson, ST; Hutchinson, J; Leveridge, MV; Nicholls, H; Smith, IED; Somers, DO; Sneddon, HF; Uddin, S; Cleasby, A; Mortenson, PN; Richardson, C; Saxty, G The discovery of quinoline-3-carboxamides as hematopoietic prostaglandin D synthase (H-PGDS) inhibitors. Bioorg Med Chem27:1456-1478 (2019) [PubMed] Article