| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Induced myeloid leukemia cell differentiation protein Mcl-1 |
|---|
| Ligand | BDBM50508940 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_2300376 |
|---|
| Kd | 3540±n/a nM |
|---|
| Citation |  Johannes, JW; Bates, S; Beigie, C; Belmonte, MA; Breen, J; Cao, S; Centrella, PA; Clark, MA; Cuozzo, JW; Dumelin, CE; Ferguson, AD; Habeshian, S; Hargreaves, D; Joubran, C; Kazmirski, S; Keefe, AD; Lamb, ML; Lan, H; Li, Y; Ma, H; Mlynarski, S; Packer, MJ; Rawlins, PB; Robbins, DW; Shen, H; Sigel, EA; Soutter, HH; Su, N; Troast, DM; Wang, H; Wickson, KF; Wu, C; Zhang, Y; Zhao, Q; Zheng, X; Hird, AW Structure Based Design of Non-Natural Peptidic Macrocyclic Mcl-1 Inhibitors. ACS Med Chem Lett8:239-244 (2017) [PubMed] Johannes, JW; Bates, S; Beigie, C; Belmonte, MA; Breen, J; Cao, S; Centrella, PA; Clark, MA; Cuozzo, JW; Dumelin, CE; Ferguson, AD; Habeshian, S; Hargreaves, D; Joubran, C; Kazmirski, S; Keefe, AD; Lamb, ML; Lan, H; Li, Y; Ma, H; Mlynarski, S; Packer, MJ; Rawlins, PB; Robbins, DW; Shen, H; Sigel, EA; Soutter, HH; Su, N; Troast, DM; Wang, H; Wickson, KF; Wu, C; Zhang, Y; Zhao, Q; Zheng, X; Hird, AW Structure Based Design of Non-Natural Peptidic Macrocyclic Mcl-1 Inhibitors. ACS Med Chem Lett8:239-244 (2017) [PubMed] |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Induced myeloid leukemia cell differentiation protein Mcl-1 |
|---|
| Name: | Induced myeloid leukemia cell differentiation protein Mcl-1 |
|---|
| Synonyms: | BCL2L3 | Bcl-2-like protein 3 | Bcl-2-like protein 3 (Mcl-1) | Bcl-2-related protein EAT/mcl1 | Bcl2-L-3 | Induced myeloid leukemia cell differentiation protein (Mcl-1) | MCL1 | MCL1_HUMAN | Mcl-1 | Myeloid Cell factor-1 (Mcl-1) | Myeloid cell leukemia sequence 1 (BCL2-related) | mcl1/EAT |
|---|
| Type: | Membrane; Single-pass membrane protein |
|---|
| Mol. Mass.: | 37332.87 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | Q07820 |
|---|
| Residue: | 350 |
|---|
| Sequence: | MFGLKRNAVIGLNLYCGGAGLGAGSGGATRPGGRLLATEKEASARREIGGGEAGAVIGGS
AGASPPSTLTPDSRRVARPPPIGAEVPDVTATPARLLFFAPTRRAAPLEEMEAPAADAIM
SPEEELDGYEPEPLGKRPAVLPLLELVGESGNNTSTDGSLPSTPPPAEEEEDELYRQSLE
IISRYLREQATGAKDTKPMGRSGATSRKALETLRRVGDGVQRNHETAFQGMLRKLDIKNE
DDVKSLSRVMIHVFSDGVTNWGRIVTLISFGAFVAKHLKTINQESCIEPLAESITDVLVR
TKRDWLVKQRGWDGFVEFFHVEDLEGGIRNVLLAFAGVAGVGAGLAYLIR
|
|
|
|---|
| BDBM50508940 |
|---|
| n/a |
|---|
| Name | BDBM50508940 |
|---|
| Synonyms: | CHEMBL4582512 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C37H42Cl3N5O4 |
|---|
| Mol. Mass. | 727.119 |
|---|
| SMILES | C[C@@H]1CCN(C)C(=O)C[C@@H](Cc2ccc(Cl)c(Cl)c2)NC(=O)CN2c3ccccc3CC[C@H](NC(=O)C[C@H](Cc3ccc(Cl)cc3)N1)C2=O |r| |
|---|
| Structure | 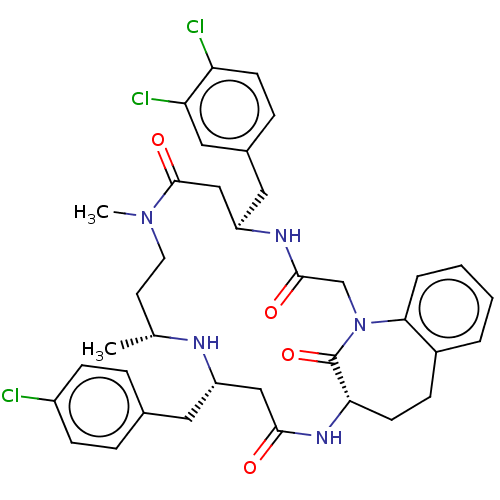
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Johannes, JW; Bates, S; Beigie, C; Belmonte, MA; Breen, J; Cao, S; Centrella, PA; Clark, MA; Cuozzo, JW; Dumelin, CE; Ferguson, AD; Habeshian, S; Hargreaves, D; Joubran, C; Kazmirski, S; Keefe, AD; Lamb, ML; Lan, H; Li, Y; Ma, H; Mlynarski, S; Packer, MJ; Rawlins, PB; Robbins, DW; Shen, H; Sigel, EA; Soutter, HH; Su, N; Troast, DM; Wang, H; Wickson, KF; Wu, C; Zhang, Y; Zhao, Q; Zheng, X; Hird, AW Structure Based Design of Non-Natural Peptidic Macrocyclic Mcl-1 Inhibitors. ACS Med Chem Lett8:239-244 (2017) [PubMed]
Johannes, JW; Bates, S; Beigie, C; Belmonte, MA; Breen, J; Cao, S; Centrella, PA; Clark, MA; Cuozzo, JW; Dumelin, CE; Ferguson, AD; Habeshian, S; Hargreaves, D; Joubran, C; Kazmirski, S; Keefe, AD; Lamb, ML; Lan, H; Li, Y; Ma, H; Mlynarski, S; Packer, MJ; Rawlins, PB; Robbins, DW; Shen, H; Sigel, EA; Soutter, HH; Su, N; Troast, DM; Wang, H; Wickson, KF; Wu, C; Zhang, Y; Zhao, Q; Zheng, X; Hird, AW Structure Based Design of Non-Natural Peptidic Macrocyclic Mcl-1 Inhibitors. ACS Med Chem Lett8:239-244 (2017) [PubMed]