| Reaction Details |
|---|
 | Report a problem with these data |
| Target | Nuclear receptor subfamily 1 group I member 2 |
|---|
| Ligand | BDBM50385834 |
|---|
| Substrate/Competitor | n/a |
|---|
| Meas. Tech. | ChEMBL_823584 (CHEMBL2044585) |
|---|
| IC50 | >50000±n/a nM |
|---|
| Citation |  Vachal, P; Miao, S; Pierce, JM; Guiadeen, D; Colandrea, VJ; Wyvratt, MJ; Salowe, SP; Sonatore, LM; Milligan, JA; Hajdu, R; Gollapudi, A; Keohane, CA; Lingham, RB; Mandala, SM; DeMartino, JA; Tong, X; Wolff, M; Steinhuebel, D; Kieczykowski, GR; Fleitz, FJ; Chapman, K; Athanasopoulos, J; Adam, G; Akyuz, CD; Jena, DK; Lusen, JW; Meng, J; Stein, BD; Xia, L; Sherer, EC; Hale, JJ 1,3,8-Triazaspiro[4.5]decane-2,4-diones as efficacious pan-inhibitors of hypoxia-inducible factor prolyl hydroxylase 1-3 (HIF PHD1-3) for the treatment of anemia. J Med Chem55:2945-59 (2012) [PubMed] Article Vachal, P; Miao, S; Pierce, JM; Guiadeen, D; Colandrea, VJ; Wyvratt, MJ; Salowe, SP; Sonatore, LM; Milligan, JA; Hajdu, R; Gollapudi, A; Keohane, CA; Lingham, RB; Mandala, SM; DeMartino, JA; Tong, X; Wolff, M; Steinhuebel, D; Kieczykowski, GR; Fleitz, FJ; Chapman, K; Athanasopoulos, J; Adam, G; Akyuz, CD; Jena, DK; Lusen, JW; Meng, J; Stein, BD; Xia, L; Sherer, EC; Hale, JJ 1,3,8-Triazaspiro[4.5]decane-2,4-diones as efficacious pan-inhibitors of hypoxia-inducible factor prolyl hydroxylase 1-3 (HIF PHD1-3) for the treatment of anemia. J Med Chem55:2945-59 (2012) [PubMed] Article |
|---|
| More Info.: | Get all data from this article, Assay Method |
|---|
| |
| Nuclear receptor subfamily 1 group I member 2 |
|---|
| Name: | Nuclear receptor subfamily 1 group I member 2 |
|---|
| Synonyms: | NR1I2 | NR1I2_HUMAN | Orphan nuclear receptor PAR1 | Orphan nuclear receptor PXR | PXR | Pregnane X receptor | SXR | Steroid and xenobiotic receptor | nuclear receptor subfamily 1 group I member 2 isoform 1 |
|---|
| Type: | Nuclear receptor |
|---|
| Mol. Mass.: | 49774.77 |
|---|
| Organism: | Homo sapiens (Human) |
|---|
| Description: | O75469 |
|---|
| Residue: | 434 |
|---|
| Sequence: | MEVRPKESWNHADFVHCEDTESVPGKPSVNADEEVGGPQICRVCGDKATGYHFNVMTCEG
CKGFFRRAMKRNARLRCPFRKGACEITRKTRRQCQACRLRKCLESGMKKEMIMSDEAVEE
RRALIKRKKSERTGTQPLGVQGLTEEQRMMIRELMDAQMKTFDTTFSHFKNFRLPGVLSS
GCELPESLQAPSREEAAKWSQVRKDLCSLKVSLQLRGEDGSVWNYKPPADSGGKEIFSLL
PHMADMSTYMFKGIISFAKVISYFRDLPIEDQISLLKGAAFELCQLRFNTVFNAETGTWE
CGRLSYCLEDTAGGFQQLLLEPMLKFHYMLKKLQLHEEEYVLMQAISLFSPDRPGVLQHR
VVDQLQEQFAITLKSYIECNRPQPAHRFLFLKIMAMLTELRSINAQHTQRLLRIQDIHPF
ATPLMQELFGITGS
|
|
|
|---|
| BDBM50385834 |
|---|
| n/a |
|---|
| Name | BDBM50385834 |
|---|
| Synonyms: | CHEMBL2043006 |
|---|
| Type | Small organic molecule |
|---|
| Emp. Form. | C31H29N5O3 |
|---|
| Mol. Mass. | 519.5937 |
|---|
| SMILES | Cc1cccnc1CN1CCC2(CC1)N(C(=O)N(C2=O)c1ccc(cc1)-c1ccccc1)c1cc(O)ccn1 |
|---|
| Structure | 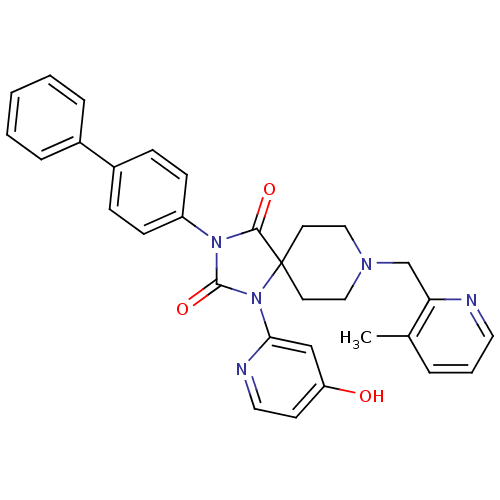
|
|---|


 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data

 Vachal, P; Miao, S; Pierce, JM; Guiadeen, D; Colandrea, VJ; Wyvratt, MJ; Salowe, SP; Sonatore, LM; Milligan, JA; Hajdu, R; Gollapudi, A; Keohane, CA; Lingham, RB; Mandala, SM; DeMartino, JA; Tong, X; Wolff, M; Steinhuebel, D; Kieczykowski, GR; Fleitz, FJ; Chapman, K; Athanasopoulos, J; Adam, G; Akyuz, CD; Jena, DK; Lusen, JW; Meng, J; Stein, BD; Xia, L; Sherer, EC; Hale, JJ 1,3,8-Triazaspiro[4.5]decane-2,4-diones as efficacious pan-inhibitors of hypoxia-inducible factor prolyl hydroxylase 1-3 (HIF PHD1-3) for the treatment of anemia. J Med Chem55:2945-59 (2012) [PubMed] Article
Vachal, P; Miao, S; Pierce, JM; Guiadeen, D; Colandrea, VJ; Wyvratt, MJ; Salowe, SP; Sonatore, LM; Milligan, JA; Hajdu, R; Gollapudi, A; Keohane, CA; Lingham, RB; Mandala, SM; DeMartino, JA; Tong, X; Wolff, M; Steinhuebel, D; Kieczykowski, GR; Fleitz, FJ; Chapman, K; Athanasopoulos, J; Adam, G; Akyuz, CD; Jena, DK; Lusen, JW; Meng, J; Stein, BD; Xia, L; Sherer, EC; Hale, JJ 1,3,8-Triazaspiro[4.5]decane-2,4-diones as efficacious pan-inhibitors of hypoxia-inducible factor prolyl hydroxylase 1-3 (HIF PHD1-3) for the treatment of anemia. J Med Chem55:2945-59 (2012) [PubMed] Article