

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Protease (Human immunodeficiency virus 1 (HIV-1)) | BDBM50469417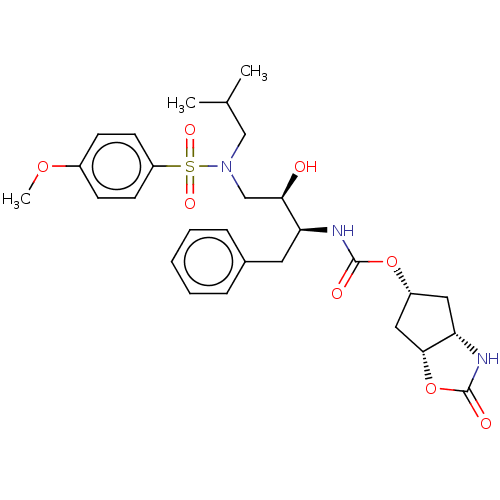 (CHEMBL4293023) | PDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid PDB UniChem | Article PubMed | 0.00120 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Purdue University Curated by ChEMBL | Assay Description Inhibition of HIV1 wild type NL4-3 protease expressed in Escherichia coli Rosetta (DE3) pLysS using Abz-Thr-Ile-Nle-Phe-(pNO2)-Gln-Arg-NH2 as substra... | J Med Chem 61: 9722-9737 (2018) Article DOI: 10.1021/acs.jmedchem.8b01227 BindingDB Entry DOI: 10.7270/Q25T3P6J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Protease (Human immunodeficiency virus 1 (HIV-1)) | BDBM50469413 (CHEMBL4286231) | PDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 0.0160 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Purdue University Curated by ChEMBL | Assay Description Inhibition of HIV1 wild type NL4-3 protease expressed in Escherichia coli Rosetta (DE3) pLysS using Abz-Thr-Ile-Nle-Phe-(pNO2)-Gln-Arg-NH2 as substra... | J Med Chem 61: 9722-9737 (2018) Article DOI: 10.1021/acs.jmedchem.8b01227 BindingDB Entry DOI: 10.7270/Q25T3P6J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Protease (Human immunodeficiency virus 1 (HIV-1)) | BDBM8125 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 0.0160 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Purdue University Curated by ChEMBL | Assay Description Inhibition of HIV1 protease using Ac-Thr-Ile-Nle-Nle-Gln-Arg-NH2 substrate by continuous fluorometric assay | Bioorg Med Chem Lett 29: 2565-2570 (2019) Article DOI: 10.1016/j.bmcl.2019.08.006 BindingDB Entry DOI: 10.7270/Q2JM2F1C | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Protease (Human immunodeficiency virus 1 (HIV-1)) | BDBM50469407 (CHEMBL4286714) | PDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 0.0160 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Purdue University Curated by ChEMBL | Assay Description Inhibition of HIV1 wild type NL4-3 protease expressed in Escherichia coli Rosetta (DE3) pLysS using Abz-Thr-Ile-Nle-Phe-(pNO2)-Gln-Arg-NH2 as substra... | J Med Chem 61: 9722-9737 (2018) Article DOI: 10.1021/acs.jmedchem.8b01227 BindingDB Entry DOI: 10.7270/Q25T3P6J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Protease (Human immunodeficiency virus 1 (HIV-1)) | BDBM8125 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 0.0160 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Purdue University Curated by ChEMBL | Assay Description Inhibition of HIV1 wild type NL4-3 protease expressed in Escherichia coli Rosetta (DE3) pLysS using Abz-Thr-Ile-Nle-Phe-(pNO2)-Gln-Arg-NH2 as substra... | J Med Chem 61: 9722-9737 (2018) Article DOI: 10.1021/acs.jmedchem.8b01227 BindingDB Entry DOI: 10.7270/Q25T3P6J | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Protease (Human immunodeficiency virus 1 (HIV-1)) | BDBM50469410 (CHEMBL4277886) | PDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 0.0300 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Purdue University Curated by ChEMBL | Assay Description Inhibition of HIV1 wild type NL4-3 protease expressed in Escherichia coli Rosetta (DE3) pLysS using Abz-Thr-Ile-Nle-Phe-(pNO2)-Gln-Arg-NH2 as substra... | J Med Chem 61: 9722-9737 (2018) Article DOI: 10.1021/acs.jmedchem.8b01227 BindingDB Entry DOI: 10.7270/Q25T3P6J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Protease (Human immunodeficiency virus 1 (HIV-1)) | BDBM50469405 (CHEMBL4292006) | PDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 0.0400 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Purdue University Curated by ChEMBL | Assay Description Inhibition of HIV1 wild type NL4-3 protease expressed in Escherichia coli Rosetta (DE3) pLysS using Abz-Thr-Ile-Nle-Phe-(pNO2)-Gln-Arg-NH2 as substra... | J Med Chem 61: 9722-9737 (2018) Article DOI: 10.1021/acs.jmedchem.8b01227 BindingDB Entry DOI: 10.7270/Q25T3P6J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| 15-hydroxyprostaglandin dehydrogenase [NAD(+)] (Homo sapiens (Human)) | BDBM50266122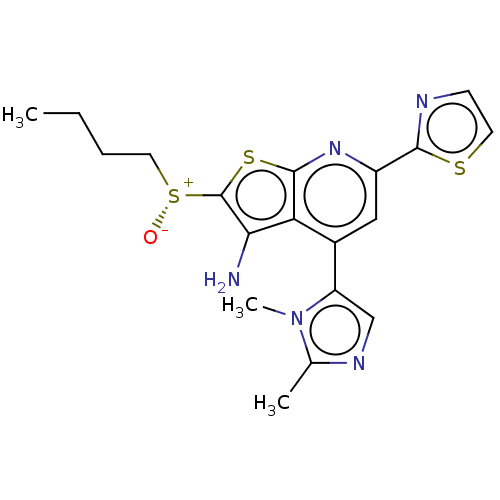 (CHEMBL4070789) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 0.0600 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
UT Southwestern Medical Center Curated by ChEMBL | Assay Description Inhibition of recombinant human C-terminal 6xHis-tagged 15-PGDH expressed in Escherichia coli using PGE2 as substrate after 15 mins in presence of NA... | J Med Chem 60: 3979-4001 (2017) Article DOI: 10.1021/acs.jmedchem.7b00271 BindingDB Entry DOI: 10.7270/Q2280B21 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Protease (Human immunodeficiency virus 1 (HIV-1)) | BDBM50469412 (CHEMBL4277970) | PDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 0.0800 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Purdue University Curated by ChEMBL | Assay Description Inhibition of HIV1 wild type NL4-3 protease expressed in Escherichia coli Rosetta (DE3) pLysS using Abz-Thr-Ile-Nle-Phe-(pNO2)-Gln-Arg-NH2 as substra... | J Med Chem 61: 9722-9737 (2018) Article DOI: 10.1021/acs.jmedchem.8b01227 BindingDB Entry DOI: 10.7270/Q25T3P6J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| 15-hydroxyprostaglandin dehydrogenase [NAD(+)] (Homo sapiens (Human)) | BDBM50266252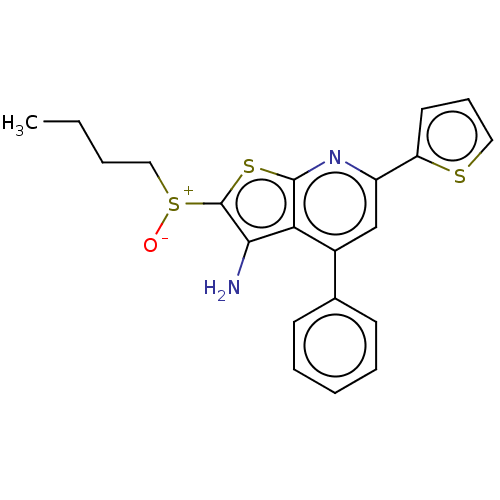 (CHEMBL4061483) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 0.100 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
TBA | Citation and Details Article DOI: 10.1021/acs.jmedchem.2c01299 BindingDB Entry DOI: 10.7270/Q23X8BSB | ||||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| 15-hydroxyprostaglandin dehydrogenase [NAD(+)] (Homo sapiens (Human)) | BDBM50266252 (CHEMBL4061483) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD DrugBank antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 0.100 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
UT Southwestern Medical Center Curated by ChEMBL | Assay Description Inhibition of 15-PGDH (unknown origin) using PGE2 as substrate preincubated for 12 hrs followed by dialysis for 12 hrs and subsequent addition of NAD... | J Med Chem 60: 3979-4001 (2017) Article DOI: 10.1021/acs.jmedchem.7b00271 BindingDB Entry DOI: 10.7270/Q2280B21 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Protease (Human immunodeficiency virus 1 (HIV-1)) | BDBM50469418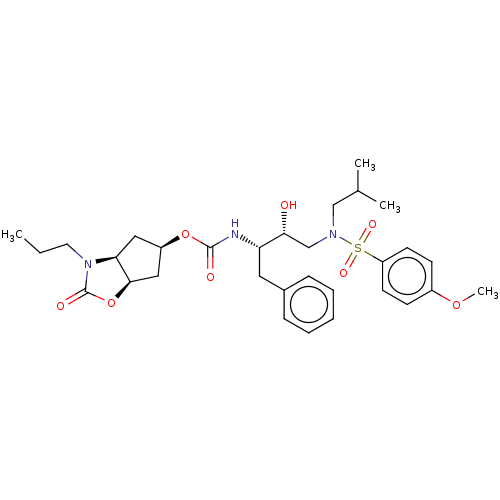 (CHEMBL4283819) | PDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 0.140 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Purdue University Curated by ChEMBL | Assay Description Inhibition of HIV1 wild type NL4-3 protease expressed in Escherichia coli Rosetta (DE3) pLysS using Abz-Thr-Ile-Nle-Phe-(pNO2)-Gln-Arg-NH2 as substra... | J Med Chem 61: 9722-9737 (2018) Article DOI: 10.1021/acs.jmedchem.8b01227 BindingDB Entry DOI: 10.7270/Q25T3P6J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Protease (Human immunodeficiency virus 1 (HIV-1)) | BDBM50469406 (CHEMBL4277539) | PDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 0.160 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Purdue University Curated by ChEMBL | Assay Description Inhibition of HIV1 wild type NL4-3 protease expressed in Escherichia coli Rosetta (DE3) pLysS using Abz-Thr-Ile-Nle-Phe-(pNO2)-Gln-Arg-NH2 as substra... | J Med Chem 61: 9722-9737 (2018) Article DOI: 10.1021/acs.jmedchem.8b01227 BindingDB Entry DOI: 10.7270/Q25T3P6J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Protease (Human immunodeficiency virus 1 (HIV-1)) | BDBM50469409 (CHEMBL4278306) | PDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 0.160 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Purdue University Curated by ChEMBL | Assay Description Inhibition of HIV1 wild type NL4-3 protease expressed in Escherichia coli Rosetta (DE3) pLysS using Abz-Thr-Ile-Nle-Phe-(pNO2)-Gln-Arg-NH2 as substra... | J Med Chem 61: 9722-9737 (2018) Article DOI: 10.1021/acs.jmedchem.8b01227 BindingDB Entry DOI: 10.7270/Q25T3P6J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Protease (Human immunodeficiency virus 1 (HIV-1)) | BDBM50469408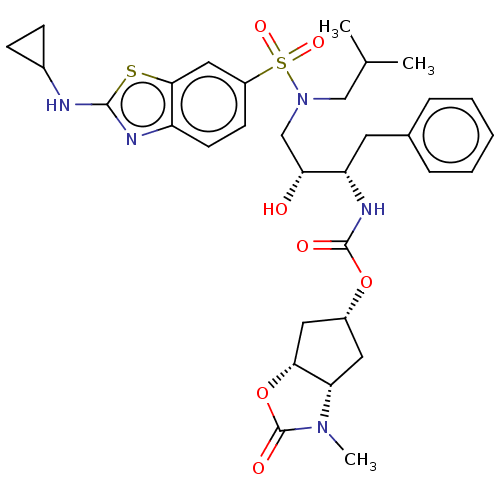 (CHEMBL4278989) | PDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 0.190 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Purdue University Curated by ChEMBL | Assay Description Inhibition of HIV1 wild type NL4-3 protease expressed in Escherichia coli Rosetta (DE3) pLysS using Abz-Thr-Ile-Nle-Phe-(pNO2)-Gln-Arg-NH2 as substra... | J Med Chem 61: 9722-9737 (2018) Article DOI: 10.1021/acs.jmedchem.8b01227 BindingDB Entry DOI: 10.7270/Q25T3P6J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Protease (Human immunodeficiency virus 1 (HIV-1)) | BDBM50469416 (CHEMBL4286215) | PDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid PDB UniChem | Article PubMed | 0.330 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Purdue University Curated by ChEMBL | Assay Description Inhibition of HIV1 wild type NL4-3 protease expressed in Escherichia coli Rosetta (DE3) pLysS using Abz-Thr-Ile-Nle-Phe-(pNO2)-Gln-Arg-NH2 as substra... | J Med Chem 61: 9722-9737 (2018) Article DOI: 10.1021/acs.jmedchem.8b01227 BindingDB Entry DOI: 10.7270/Q25T3P6J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Protease (Human immunodeficiency virus 1 (HIV-1)) | BDBM50522184 (CHEMBL4444017) | PDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 0.510 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Purdue University Curated by ChEMBL | Assay Description Inhibition of HIV1 protease using Ac-Thr-Ile-Nle-Nle-Gln-Arg-NH2 substrate by continuous fluorometric assay | Bioorg Med Chem Lett 29: 2565-2570 (2019) Article DOI: 10.1016/j.bmcl.2019.08.006 BindingDB Entry DOI: 10.7270/Q2JM2F1C | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Protease (Human immunodeficiency virus 1 (HIV-1)) | BDBM50469411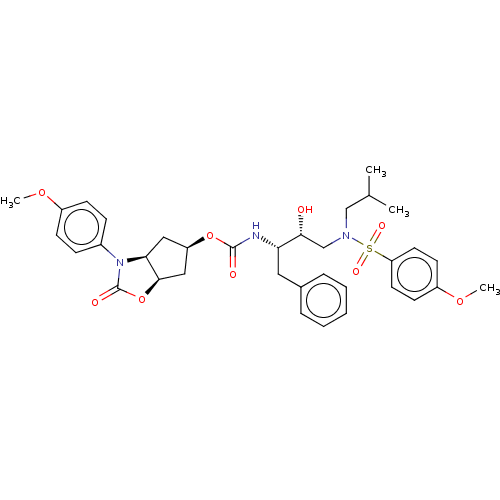 (CHEMBL4279922) | PDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 0.630 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Purdue University Curated by ChEMBL | Assay Description Inhibition of HIV1 wild type NL4-3 protease expressed in Escherichia coli Rosetta (DE3) pLysS using Abz-Thr-Ile-Nle-Phe-(pNO2)-Gln-Arg-NH2 as substra... | J Med Chem 61: 9722-9737 (2018) Article DOI: 10.1021/acs.jmedchem.8b01227 BindingDB Entry DOI: 10.7270/Q25T3P6J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Delta-type opioid receptor (Rattus norvegicus (rat)) | BDBM50001092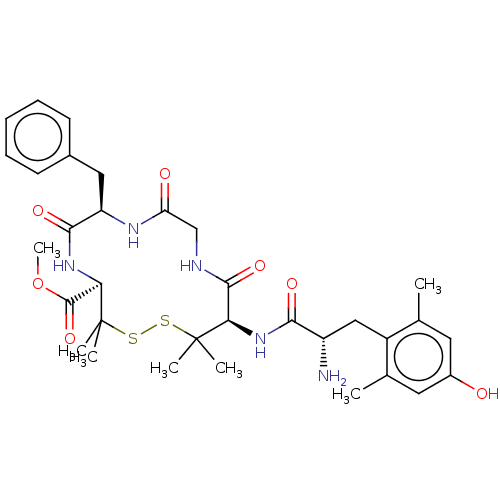 (13-[2-Amino-3-(4-hydroxy-2,6-dimethyl-phenyl)-prop...) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | 0.900 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
G.D. Searle and Company Curated by ChEMBL | Assay Description Compound was evaluated for its binding affinity by displacement [3H]- DSLET from Opioid receptor delta 1 by using opioid radioligand binding assay | J Med Chem 35: 2928-38 (1992) BindingDB Entry DOI: 10.7270/Q2ST7QF6 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Mu-type opioid receptor (Rattus norvegicus (rat)) | BDBM50001092 (13-[2-Amino-3-(4-hydroxy-2,6-dimethyl-phenyl)-prop...) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | 0.900 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
G.D. Searle and Company Curated by ChEMBL | Assay Description Compound was evaluated for its binding affinity by displacement of [3H]- DAMPGO from opioid receptor mu by using opioid radioligand binding assay | J Med Chem 35: 2928-38 (1992) BindingDB Entry DOI: 10.7270/Q2ST7QF6 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Protease (Human immunodeficiency virus 1 (HIV-1)) | BDBM50469414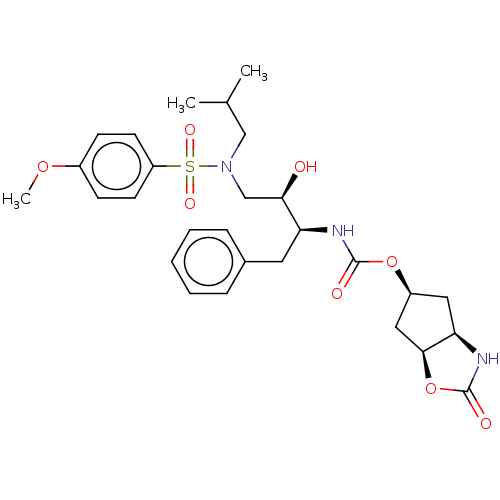 (CHEMBL4288541) | PDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 1.30 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Purdue University Curated by ChEMBL | Assay Description Inhibition of HIV1 wild type NL4-3 protease expressed in Escherichia coli Rosetta (DE3) pLysS using Abz-Thr-Ile-Nle-Phe-(pNO2)-Gln-Arg-NH2 as substra... | J Med Chem 61: 9722-9737 (2018) Article DOI: 10.1021/acs.jmedchem.8b01227 BindingDB Entry DOI: 10.7270/Q25T3P6J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Delta-type opioid receptor (Rattus norvegicus (rat)) | BDBM50001090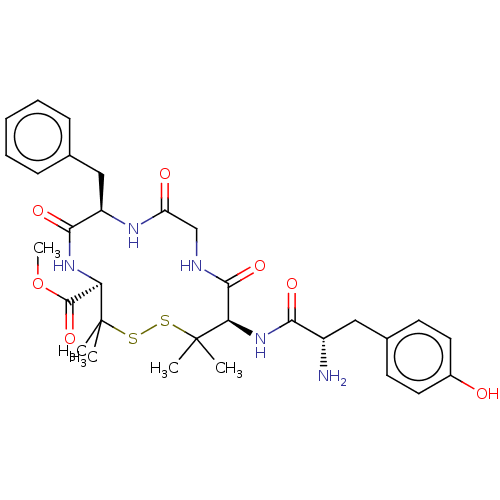 (13-[2-Amino-3-(4-hydroxy-phenyl)-propionylamino]-7...) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | 2.5 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
G.D. Searle and Company Curated by ChEMBL | Assay Description Compound was evaluated for its binding affinity by displacement [3H]- DSLET from Opioid receptor delta 1 by using opioid radioligand binding assay | J Med Chem 35: 2928-38 (1992) BindingDB Entry DOI: 10.7270/Q2ST7QF6 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Histamine H4 receptor (Rattus norvegicus (rat)) | BDBM22566 (5-chloro-2-[(4-methylpiperazin-1-yl)carbonyl]-1H-i...) | KEGG UniProtKB/SwissProt GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid UniChem Patents Similars | Article PubMed | 2.60 | -49.0 | n/a | n/a | n/a | n/a | n/a | 7.5 | 25 |
Johnson & Johnson Pharmaceutical | Assay Description The Ki values were calculated based on an experimentally determined appropriate Kd value according to Cheng and Prusoff. | J Pharmacol Exp Ther 309: 404-13 (2004) Article DOI: 10.1124/jpet.103.061754 BindingDB Entry DOI: 10.7270/Q2VX0DTD | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Histamine H4 receptor (Homo sapiens (Human)) | BDBM22566 (5-chloro-2-[(4-methylpiperazin-1-yl)carbonyl]-1H-i...) | PDB Reactome pathway KEGG UniProtKB/SwissProt DrugBank antibodypedia GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid UniChem Patents Similars | Article PubMed | 4.10 | -47.9 | n/a | n/a | n/a | n/a | n/a | 7.5 | 25 |
Johnson & Johnson Pharmaceutical | Assay Description The Ki values were calculated based on an experimentally determined appropriate Kd value according to Cheng and Prusoff. | J Pharmacol Exp Ther 309: 404-13 (2004) Article DOI: 10.1124/jpet.103.061754 BindingDB Entry DOI: 10.7270/Q2VX0DTD | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Protease (Human immunodeficiency virus 1 (HIV-1)) | BDBM50522181 (CHEMBL4456725) | PDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 4.5 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Purdue University Curated by ChEMBL | Assay Description Inhibition of HIV1 protease using Ac-Thr-Ile-Nle-Nle-Gln-Arg-NH2 substrate by continuous fluorometric assay | Bioorg Med Chem Lett 29: 2565-2570 (2019) Article DOI: 10.1016/j.bmcl.2019.08.006 BindingDB Entry DOI: 10.7270/Q2JM2F1C | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Histamine H4 receptor (Mus musculus (mouse)) | BDBM22566 (5-chloro-2-[(4-methylpiperazin-1-yl)carbonyl]-1H-i...) | Reactome pathway KEGG UniProtKB/SwissProt GoogleScholar AffyNet | Purchase CHEMBL MCE PC cid PC sid UniChem Patents Similars | Article PubMed | 4.60 | -47.6 | n/a | n/a | n/a | n/a | n/a | 7.5 | 25 |
Johnson & Johnson Pharmaceutical | Assay Description The Ki values were calculated based on an experimentally determined appropriate Kd value according to Cheng and Prusoff. | J Pharmacol Exp Ther 309: 404-13 (2004) Article DOI: 10.1124/jpet.103.061754 BindingDB Entry DOI: 10.7270/Q2VX0DTD | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Protease (Human immunodeficiency virus 1 (HIV-1)) | BDBM50522183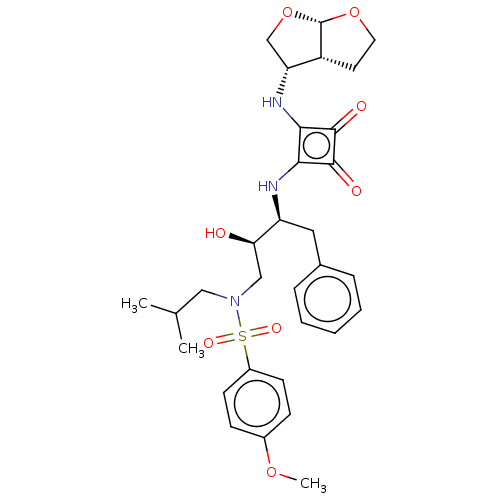 (CHEMBL4467544) | PDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 5.60 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Purdue University Curated by ChEMBL | Assay Description Inhibition of HIV1 protease using Ac-Thr-Ile-Nle-Nle-Gln-Arg-NH2 substrate by continuous fluorometric assay | Bioorg Med Chem Lett 29: 2565-2570 (2019) Article DOI: 10.1016/j.bmcl.2019.08.006 BindingDB Entry DOI: 10.7270/Q2JM2F1C | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Delta-type opioid receptor (Rattus norvegicus (rat)) | BDBM50001088 ((DPDPE)13-[2-Amino-3-(4-hydroxy-phenyl)-propionyla...) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | 7 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
G.D. Searle and Company Curated by ChEMBL | Assay Description Compound was evaluated for its binding affinity by displacement [3H]- DSLET from Opioid receptor delta 1 by using opioid radioligand binding assay | J Med Chem 35: 2928-38 (1992) BindingDB Entry DOI: 10.7270/Q2ST7QF6 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Mu-type opioid receptor (Rattus norvegicus (rat)) | BDBM50001090 (13-[2-Amino-3-(4-hydroxy-phenyl)-propionylamino]-7...) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | 9.5 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
G.D. Searle and Company Curated by ChEMBL | Assay Description Compound was evaluated for its binding affinity by displacement of [3H]- DAMPGO from opioid receptor mu by using opioid radioligand binding assay | J Med Chem 35: 2928-38 (1992) BindingDB Entry DOI: 10.7270/Q2ST7QF6 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Free fatty acid receptor 1 (Homo sapiens (Human)) | BDBM50267028 (CHEMBL4073525) | PDB UniProtKB/SwissProt antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 10 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Bristol-Myers Squibb Curated by ChEMBL | Assay Description Agonist activity at human GPR40 expressed in CHO-A12 cells assessed as increase in intracellular calcium flux measured for 100 secs by FLIPR assay | J Med Chem 60: 1417-1431 (2017) Article DOI: 10.1021/acs.jmedchem.6b01559 BindingDB Entry DOI: 10.7270/Q25X2CD5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Mu-type opioid receptor (Rattus norvegicus (rat)) | BDBM50001093 (13-[2-Amino-3-(4-hydroxy-2,6-dimethyl-phenyl)-prop...) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | 11 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
G.D. Searle and Company Curated by ChEMBL | Assay Description Compound was evaluated for its binding affinity by displacement of [3H]- DAMPGO from opioid receptor mu by using opioid radioligand binding assay | J Med Chem 35: 2928-38 (1992) BindingDB Entry DOI: 10.7270/Q2ST7QF6 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Mu-type opioid receptor (Rattus norvegicus (rat)) | BDBM50001093 (13-[2-Amino-3-(4-hydroxy-2,6-dimethyl-phenyl)-prop...) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | 11 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
G.D. Searle and Company Curated by ChEMBL | Assay Description Compound was evaluated for its binding affinity by displacement [3H]- DSLET from Opioid receptor delta 1 by using opioid radioligand binding assay | J Med Chem 35: 2928-38 (1992) BindingDB Entry DOI: 10.7270/Q2ST7QF6 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Free fatty acid receptor 1 (Homo sapiens (Human)) | BDBM50267031 (CHEMBL4079930) | PDB UniProtKB/SwissProt antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 13 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Bristol-Myers Squibb Curated by ChEMBL | Assay Description Agonist activity at human GPR40 expressed in CHO-A12 cells assessed as increase in intracellular calcium flux measured for 100 secs by FLIPR assay | J Med Chem 60: 1417-1431 (2017) Article DOI: 10.1021/acs.jmedchem.6b01559 BindingDB Entry DOI: 10.7270/Q25X2CD5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Delta-type opioid receptor (Rattus norvegicus (rat)) | BDBM50001093 (13-[2-Amino-3-(4-hydroxy-2,6-dimethyl-phenyl)-prop...) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | 21 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
G.D. Searle and Company Curated by ChEMBL | Assay Description Compound was evaluated for its binding affinity by displacement [3H]- DSLET from Opioid receptor delta 1 by using opioid radioligand binding assay | J Med Chem 35: 2928-38 (1992) BindingDB Entry DOI: 10.7270/Q2ST7QF6 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Delta-type opioid receptor (Rattus norvegicus (rat)) | BDBM50001093 (13-[2-Amino-3-(4-hydroxy-2,6-dimethyl-phenyl)-prop...) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | 21 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
G.D. Searle and Company Curated by ChEMBL | Assay Description Compound was evaluated for its binding affinity by displacement [3H]- DSLET from Opioid receptor delta 1 by using opioid radioligand binding assay | J Med Chem 35: 2928-38 (1992) BindingDB Entry DOI: 10.7270/Q2ST7QF6 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Protease (Human immunodeficiency virus 1 (HIV-1)) | BDBM50522186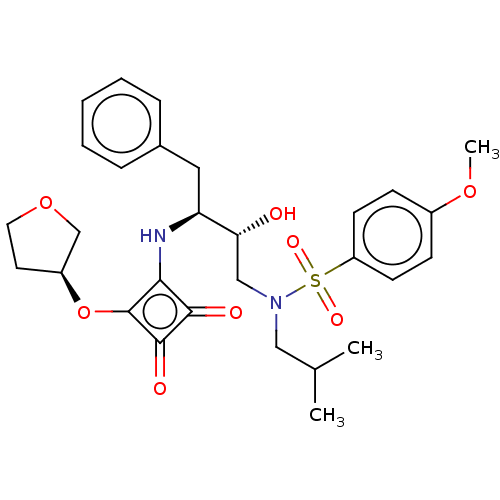 (CHEMBL4445644) | PDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 31 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Purdue University Curated by ChEMBL | Assay Description Inhibition of HIV1 protease using Ac-Thr-Ile-Nle-Nle-Gln-Arg-NH2 substrate by continuous fluorometric assay | Bioorg Med Chem Lett 29: 2565-2570 (2019) Article DOI: 10.1016/j.bmcl.2019.08.006 BindingDB Entry DOI: 10.7270/Q2JM2F1C | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Mu-type opioid receptor (Rattus norvegicus (rat)) | BDBM50001089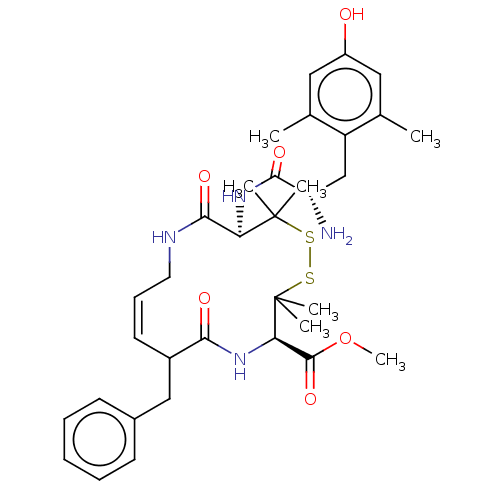 (13-[2-Amino-3-(4-hydroxy-2,6-dimethyl-phenyl)-prop...) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | 32 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
G.D. Searle and Company Curated by ChEMBL | Assay Description Dissociation rate constant of compound for mutant T46S Escherichia coli dihydrofolate reductase | J Med Chem 35: 2928-38 (1992) BindingDB Entry DOI: 10.7270/Q2ST7QF6 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Mu-type opioid receptor (Rattus norvegicus (rat)) | BDBM50001089 (13-[2-Amino-3-(4-hydroxy-2,6-dimethyl-phenyl)-prop...) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | 32 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
G.D. Searle and Company Curated by ChEMBL | Assay Description Compound was evaluated for its binding affinity by displacement of [3H]- DAMPGO from opioid receptor mu by using opioid radioligand binding assay | J Med Chem 35: 2928-38 (1992) BindingDB Entry DOI: 10.7270/Q2ST7QF6 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Free fatty acid receptor 1 (Homo sapiens (Human)) | BDBM50267009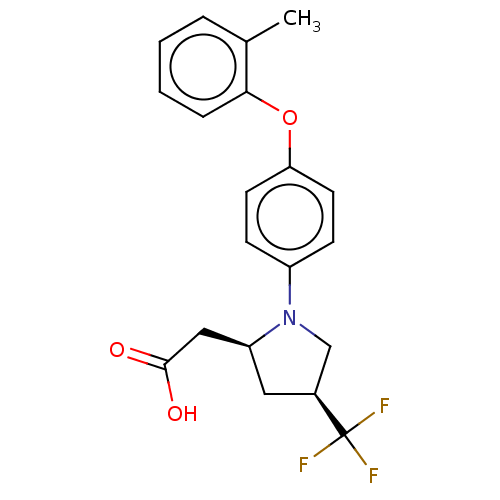 (CHEMBL4089171) | PDB UniProtKB/SwissProt antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 35 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Bristol-Myers Squibb Curated by ChEMBL | Assay Description Binding affinity to human GPR40 expressed in HEK293 cell membranes after 1 hr by radioligand displacement based scintillation counting method | J Med Chem 60: 1417-1431 (2017) Article DOI: 10.1021/acs.jmedchem.6b01559 BindingDB Entry DOI: 10.7270/Q25X2CD5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Free fatty acid receptor 1 (Homo sapiens (Human)) | BDBM50267029 (CHEMBL4069191) | PDB UniProtKB/SwissProt antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 39 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Bristol-Myers Squibb Curated by ChEMBL | Assay Description Binding affinity to human GPR40 expressed in HEK293 cell membranes after 1 hr by radioligand displacement based scintillation counting method | J Med Chem 60: 1417-1431 (2017) Article DOI: 10.1021/acs.jmedchem.6b01559 BindingDB Entry DOI: 10.7270/Q25X2CD5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Delta-type opioid receptor (Rattus norvegicus (rat)) | BDBM50001089 (13-[2-Amino-3-(4-hydroxy-2,6-dimethyl-phenyl)-prop...) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | 45 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
G.D. Searle and Company Curated by ChEMBL | Assay Description Compound was evaluated for its binding affinity by displacement [3H]- DSLET from Opioid receptor delta 1 by using opioid radioligand binding assay | J Med Chem 35: 2928-38 (1992) BindingDB Entry DOI: 10.7270/Q2ST7QF6 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Free fatty acid receptor 1 (Homo sapiens (Human)) | BDBM50267010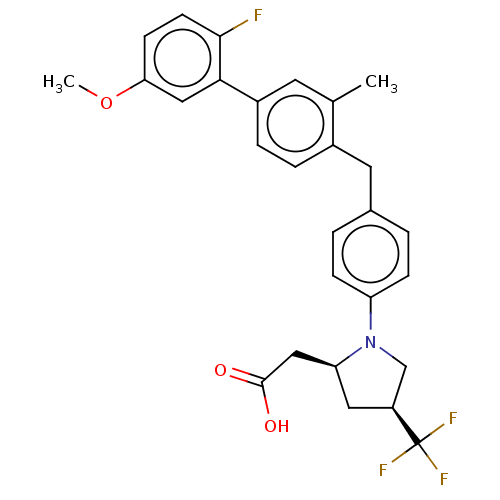 (CHEMBL4082395) | PDB UniProtKB/SwissProt antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 45 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Bristol-Myers Squibb Curated by ChEMBL | Assay Description Binding affinity to human GPR40 expressed in HEK293 cell membranes after 1 hr by radioligand displacement based scintillation counting method | J Med Chem 60: 1417-1431 (2017) Article DOI: 10.1021/acs.jmedchem.6b01559 BindingDB Entry DOI: 10.7270/Q25X2CD5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Delta-type opioid receptor (Rattus norvegicus (rat)) | BDBM50001089 (13-[2-Amino-3-(4-hydroxy-2,6-dimethyl-phenyl)-prop...) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | 45 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
G.D. Searle and Company Curated by ChEMBL | Assay Description Compound was evaluated for its binding affinity by displacement [3H]- DSLET from Opioid receptor delta 1 by using opioid radioligand binding assay | J Med Chem 35: 2928-38 (1992) BindingDB Entry DOI: 10.7270/Q2ST7QF6 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Free fatty acid receptor 1 (Homo sapiens (Human)) | BDBM50267117 (CHEMBL4060499) | PDB UniProtKB/SwissProt antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 49 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Bristol-Myers Squibb Curated by ChEMBL | Assay Description Binding affinity to human GPR40 expressed in HEK293 cell membranes after 1 hr by radioligand displacement based scintillation counting method | J Med Chem 60: 1417-1431 (2017) Article DOI: 10.1021/acs.jmedchem.6b01559 BindingDB Entry DOI: 10.7270/Q25X2CD5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Protease (Human immunodeficiency virus 1 (HIV-1)) | BDBM50522189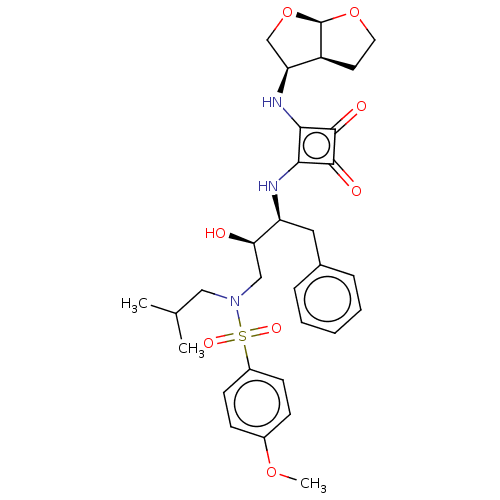 (CHEMBL4469790) | PDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 55 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Purdue University Curated by ChEMBL | Assay Description Inhibition of HIV1 protease using Ac-Thr-Ile-Nle-Nle-Gln-Arg-NH2 substrate by continuous fluorometric assay | Bioorg Med Chem Lett 29: 2565-2570 (2019) Article DOI: 10.1016/j.bmcl.2019.08.006 BindingDB Entry DOI: 10.7270/Q2JM2F1C | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Free fatty acid receptor 1 (Homo sapiens (Human)) | BDBM50267030 (CHEMBL4090240) | PDB UniProtKB/SwissProt antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 60 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Bristol-Myers Squibb Curated by ChEMBL | Assay Description Binding affinity to human GPR40 expressed in HEK293 cell membranes after 1 hr by radioligand displacement based scintillation counting method | J Med Chem 60: 1417-1431 (2017) Article DOI: 10.1021/acs.jmedchem.6b01559 BindingDB Entry DOI: 10.7270/Q25X2CD5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Free fatty acid receptor 1 (Homo sapiens (Human)) | BDBM50267044 (CHEMBL4069764) | PDB UniProtKB/SwissProt antibodypedia GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 73 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Bristol-Myers Squibb Curated by ChEMBL | Assay Description Binding affinity to human GPR40 expressed in HEK293 cell membranes after 1 hr by radioligand displacement based scintillation counting method | J Med Chem 60: 1417-1431 (2017) Article DOI: 10.1021/acs.jmedchem.6b01559 BindingDB Entry DOI: 10.7270/Q25X2CD5 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Delta-type opioid receptor (Rattus norvegicus (rat)) | BDBM50001091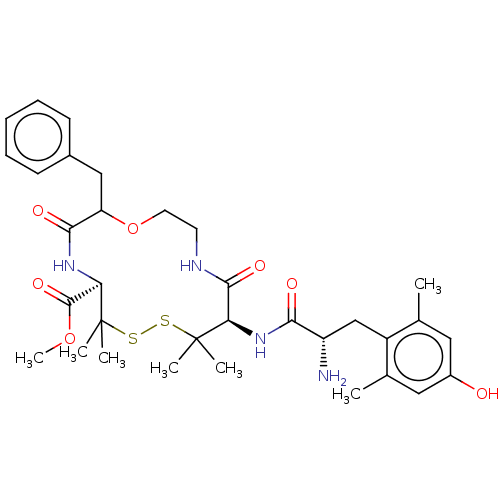 (10-[2-Amino-3-(4-hydroxy-2,6-dimethyl-phenyl)-prop...) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | 91 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
G.D. Searle and Company Curated by ChEMBL | Assay Description Compound was evaluated for its binding affinity by displacement [3H]- DSLET from Opioid receptor delta 1 by using opioid radioligand binding assay | J Med Chem 35: 2928-38 (1992) BindingDB Entry DOI: 10.7270/Q2ST7QF6 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Delta-type opioid receptor (Rattus norvegicus (rat)) | BDBM50001091 (10-[2-Amino-3-(4-hydroxy-2,6-dimethyl-phenyl)-prop...) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | PubMed | 91 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
G.D. Searle and Company Curated by ChEMBL | Assay Description Dissociation rate constant of compound for mutant T46A Escherichia coli dihydrofolate reductase | J Med Chem 35: 2928-38 (1992) BindingDB Entry DOI: 10.7270/Q2ST7QF6 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Protease (Human immunodeficiency virus 1 (HIV-1)) | BDBM50469415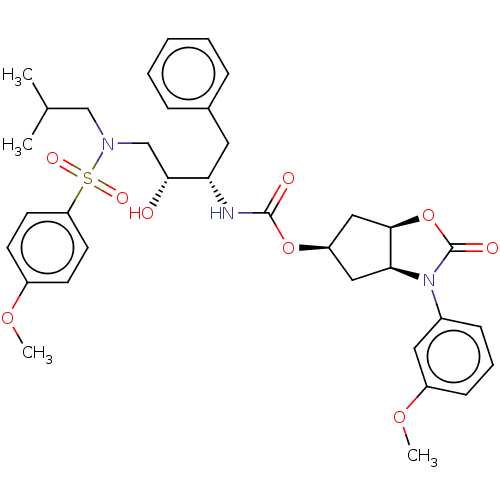 (CHEMBL4284787) | PDB UniProtKB/TrEMBL B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem | Article PubMed | 93 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Purdue University Curated by ChEMBL | Assay Description Inhibition of HIV1 wild type NL4-3 protease expressed in Escherichia coli Rosetta (DE3) pLysS using Abz-Thr-Ile-Nle-Phe-(pNO2)-Gln-Arg-NH2 as substra... | J Med Chem 61: 9722-9737 (2018) Article DOI: 10.1021/acs.jmedchem.8b01227 BindingDB Entry DOI: 10.7270/Q25T3P6J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Displayed 1 to 50 (of 1046 total ) | Next | Last >> |