

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dimer of Gag-Pol polyprotein [489-587] (Human immunodeficiency virus type 1) | BDBM4690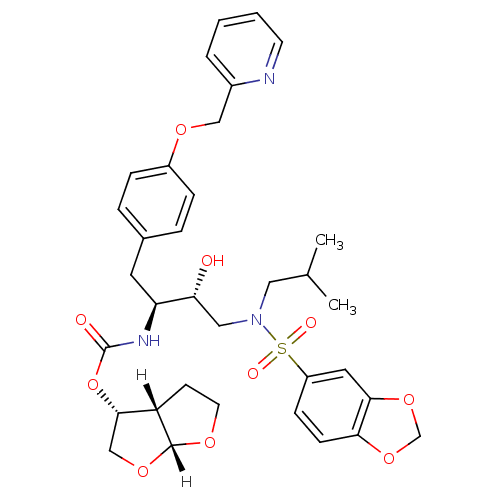 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.00000600 | -82.5 | n/a | n/a | n/a | n/a | n/a | 5.5 | 30 |
GlaxoSmithKline | Assay Description The method was a competitive displacement assay used to determine binding affinities of other inhibitors relative to that of GW0385. The inhibitor of... | Biochemistry 43: 14500-7 (2004) Article DOI: 10.1021/bi0488799 BindingDB Entry DOI: 10.7270/Q25M63WJ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [489-587] (Human immunodeficiency virus type 1) | BDBM4689 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 0.0000130 | -80.6 | n/a | n/a | n/a | n/a | n/a | 5.5 | 30 |
GlaxoSmithKline | Assay Description The method was a competitive displacement assay used to determine binding affinities of other inhibitors relative to that of GW0385. The inhibitor of... | Biochemistry 43: 14500-7 (2004) Article DOI: 10.1021/bi0488799 BindingDB Entry DOI: 10.7270/Q25M63WJ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [489-587] (Human immunodeficiency virus type 1) | BDBM4685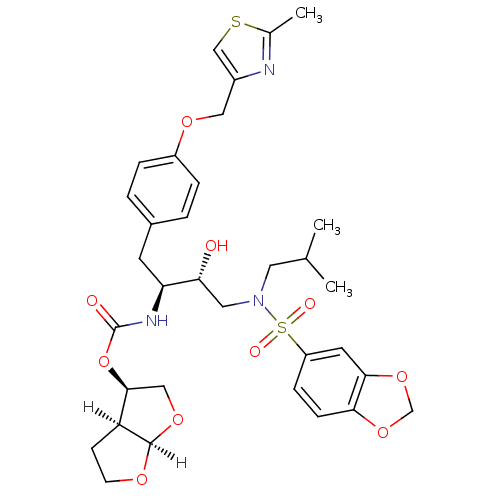 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 0.0000150 | -80.2 | n/a | n/a | n/a | n/a | n/a | 5.5 | 30 |
GlaxoSmithKline | Assay Description The assay method employed kinetic determinations of values for k1 and k-1, from which value of inhibition constant (Ki ) was determined (k-1/k1). The... | Biochemistry 43: 14500-7 (2004) Article DOI: 10.1021/bi0488799 BindingDB Entry DOI: 10.7270/Q25M63WJ | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Dimer of Gag-Pol polyprotein [489-587] (Human immunodeficiency virus type 1) | BDBM4688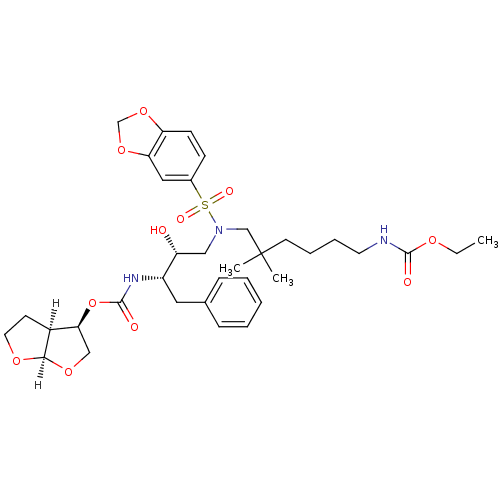 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 0.000165 | -74.2 | n/a | n/a | n/a | n/a | n/a | 5.5 | 30 |
GlaxoSmithKline | Assay Description The method was a competitive displacement assay used to determine binding affinities of other inhibitors relative to that of GW0385. The inhibitor of... | Biochemistry 43: 14500-7 (2004) Article DOI: 10.1021/bi0488799 BindingDB Entry DOI: 10.7270/Q25M63WJ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [489-587,M535I,L552P,A560V,V571F,I573V] (Human immunodeficiency virus type 1) | BDBM4689 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 0.000220 | -73.5 | n/a | n/a | n/a | n/a | n/a | 5.5 | 30 |
GlaxoSmithKline | Assay Description Enzymatic activity was determined with fluorogenic substrate in the presence and absence of inhibitor. The fluorescence increase due to hydrolysis of... | Biochemistry 43: 14500-7 (2004) Article DOI: 10.1021/bi0488799 BindingDB Entry DOI: 10.7270/Q25M63WJ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [489-587] (Human immunodeficiency virus type 1) | BDBM4687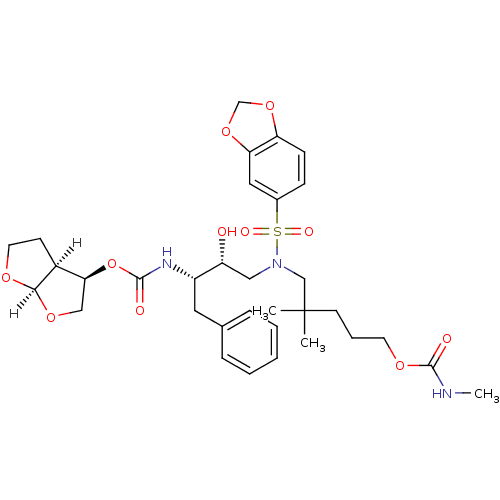 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.000240 | -73.2 | n/a | n/a | n/a | n/a | n/a | 5.5 | 30 |
GlaxoSmithKline | Assay Description The method was a competitive displacement assay used to determine binding affinities of other inhibitors relative to that of GW0385. The inhibitor of... | Biochemistry 43: 14500-7 (2004) Article DOI: 10.1021/bi0488799 BindingDB Entry DOI: 10.7270/Q25M63WJ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [489-587,M535I,L552P,A560V,V571F,I573V] (Human immunodeficiency virus type 1) | BDBM4690 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.000420 | -71.8 | n/a | n/a | n/a | n/a | n/a | 5.5 | 30 |
GlaxoSmithKline | Assay Description Enzymatic activity was determined with fluorogenic substrate in the presence and absence of inhibitor. The fluorescence increase due to hydrolysis of... | Biochemistry 43: 14500-7 (2004) Article DOI: 10.1021/bi0488799 BindingDB Entry DOI: 10.7270/Q25M63WJ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [489-587,M535I,L552P,A560V,V571F,I573V] (Human immunodeficiency virus type 1) | BDBM4685 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE MMDB PC cid PC sid PDB UniChem Patents Similars | Article PubMed | 0.000750 | -70.4 | n/a | n/a | n/a | n/a | n/a | 5.5 | 30 |
GlaxoSmithKline | Assay Description The assay method employed kinetic determinations of values for k1 and k-1, from which value of inhibition constant (Ki ) was determined (k-1/k1). The... | Biochemistry 43: 14500-7 (2004) Article DOI: 10.1021/bi0488799 BindingDB Entry DOI: 10.7270/Q25M63WJ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [489-587,L499I,L508Q,K509R,E524D,M525I,S526N,M535I,I539V,I543V,I551V,L552P,A560V,V571A,L579M] (Human immunodeficiency virus type 1) | BDBM4688 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 0.00120 | -69.2 | n/a | n/a | n/a | n/a | n/a | 5.5 | 30 |
GlaxoSmithKline | Assay Description Enzymatic activity was determined with fluorogenic substrate in the presence and absence of inhibitor. The fluorescence increase due to hydrolysis of... | Biochemistry 43: 14500-7 (2004) Article DOI: 10.1021/bi0488799 BindingDB Entry DOI: 10.7270/Q25M63WJ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [489-587,L499I,L508Q,K509R,E524D,M525I,S526N,M535I,I539V,I543V,I551V,L552P,A560V,V571A,L579M] (Human immunodeficiency virus type 1) | BDBM4690 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.00170 | -68.3 | n/a | n/a | n/a | n/a | n/a | 5.5 | 30 |
GlaxoSmithKline | Assay Description Enzymatic activity was determined with fluorogenic substrate in the presence and absence of inhibitor. The fluorescence increase due to hydrolysis of... | Biochemistry 43: 14500-7 (2004) Article DOI: 10.1021/bi0488799 BindingDB Entry DOI: 10.7270/Q25M63WJ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [489-587,I539V] (Human immunodeficiency virus type 1) | BDBM4685 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 0.00200 | -67.9 | n/a | n/a | n/a | n/a | n/a | 5.5 | 30 |
GlaxoSmithKline | Assay Description The assay method employed kinetic determinations of values for k1 and k-1, from which value of inhibition constant (Ki ) was determined (k-1/k1). The... | Biochemistry 43: 14500-7 (2004) Article DOI: 10.1021/bi0488799 BindingDB Entry DOI: 10.7270/Q25M63WJ | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Dimer of Gag-Pol polyprotein [489-587,L499I,L508Q,K509R,E524D,M525I,S526N,M535I,I539V,I543V,I551V,L552P,A560V,V571A,L579M] (Human immunodeficiency virus type 1) | BDBM4689 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 0.00240 | -67.4 | n/a | n/a | n/a | n/a | n/a | 5.5 | 30 |
GlaxoSmithKline | Assay Description Enzymatic activity was determined with fluorogenic substrate in the presence and absence of inhibitor. The fluorescence increase due to hydrolysis of... | Biochemistry 43: 14500-7 (2004) Article DOI: 10.1021/bi0488799 BindingDB Entry DOI: 10.7270/Q25M63WJ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [489-587,I539V] (Human immunodeficiency virus type 1) | BDBM4690 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.00260 | -67.2 | n/a | n/a | n/a | n/a | n/a | 5.5 | 30 |
GlaxoSmithKline | Assay Description The method was a competitive displacement assay used to determine binding affinities of other inhibitors relative to that of GW0385. The inhibitor of... | Biochemistry 43: 14500-7 (2004) Article DOI: 10.1021/bi0488799 BindingDB Entry DOI: 10.7270/Q25M63WJ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [489-587,L499I,L508Q,K509R,E524D,M525I,S526N,M535I,I539V,I543V,I551V,L552P,A560V,V571A,L579M] (Human immunodeficiency virus type 1) | BDBM4685 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE MMDB PC cid PC sid PDB UniChem Patents Similars | MMDB PDB Article PubMed | 0.00340 | -66.6 | n/a | n/a | n/a | n/a | n/a | 5.5 | 30 |
GlaxoSmithKline | Assay Description The assay method employed kinetic determinations of values for k1 and k-1, from which value of inhibition constant (Ki ) was determined (k-1/k1). The... | Biochemistry 43: 14500-7 (2004) Article DOI: 10.1021/bi0488799 BindingDB Entry DOI: 10.7270/Q25M63WJ | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Dimer of Gag-Pol polyprotein [489-587,I539V] (Human immunodeficiency virus type 1) | BDBM4689 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 0.00390 | -66.2 | n/a | n/a | n/a | n/a | n/a | 5.5 | 30 |
GlaxoSmithKline | Assay Description The method was a competitive displacement assay used to determine binding affinities of other inhibitors relative to that of GW0385. The inhibitor of... | Biochemistry 43: 14500-7 (2004) Article DOI: 10.1021/bi0488799 BindingDB Entry DOI: 10.7270/Q25M63WJ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [489-587,M535I,L552P,A560V,V571F,I573V] (Human immunodeficiency virus type 1) | BDBM4688 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 0.00430 | -66.0 | n/a | n/a | n/a | n/a | n/a | 5.5 | 30 |
GlaxoSmithKline | Assay Description Enzymatic activity was determined with fluorogenic substrate in the presence and absence of inhibitor. The fluorescence increase due to hydrolysis of... | Biochemistry 43: 14500-7 (2004) Article DOI: 10.1021/bi0488799 BindingDB Entry DOI: 10.7270/Q25M63WJ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [489-587,I539V] (Human immunodeficiency virus type 1) | BDBM4688 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | 0.00460 | -65.8 | n/a | n/a | n/a | n/a | n/a | 5.5 | 30 |
GlaxoSmithKline | Assay Description The method was a competitive displacement assay used to determine binding affinities of other inhibitors relative to that of GW0385. The inhibitor of... | Biochemistry 43: 14500-7 (2004) Article DOI: 10.1021/bi0488799 BindingDB Entry DOI: 10.7270/Q25M63WJ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [489-587,M535I,L552P,A560V,V571F,I573V] (Human immunodeficiency virus type 1) | BDBM4687 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.0250 | -61.5 | n/a | n/a | n/a | n/a | n/a | 5.5 | 30 |
GlaxoSmithKline | Assay Description Enzymatic activity was determined with fluorogenic substrate in the presence and absence of inhibitor. The fluorescence increase due to hydrolysis of... | Biochemistry 43: 14500-7 (2004) Article DOI: 10.1021/bi0488799 BindingDB Entry DOI: 10.7270/Q25M63WJ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [489-587,I539V] (Human immunodeficiency virus type 1) | BDBM4687 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.0270 | -61.3 | n/a | n/a | n/a | n/a | n/a | 5.5 | 30 |
GlaxoSmithKline | Assay Description The method was a competitive displacement assay used to determine binding affinities of other inhibitors relative to that of GW0385. The inhibitor of... | Biochemistry 43: 14500-7 (2004) Article DOI: 10.1021/bi0488799 BindingDB Entry DOI: 10.7270/Q25M63WJ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [489-587,L499I,L508Q,K509R,E524D,M525I,S526N,M535I,I539V,I543V,I551V,L552P,A560V,V571A,L579M] (Human immunodeficiency virus type 1) | BDBM4687 ((3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl N-[(2S...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | PC cid PC sid UniChem Patents Similars | Article PubMed | 0.0544 | -59.6 | n/a | n/a | n/a | n/a | n/a | 5.5 | 30 |
GlaxoSmithKline | Assay Description Enzymatic activity was determined with fluorogenic substrate in the presence and absence of inhibitor. The fluorescence increase due to hydrolysis of... | Biochemistry 43: 14500-7 (2004) Article DOI: 10.1021/bi0488799 BindingDB Entry DOI: 10.7270/Q25M63WJ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [489-587] (Human immunodeficiency virus type 1) | BDBM577 ((3S)-oxolan-3-yl N-[(2S,3R)-4-[(4-aminobenzene)(2-...) | PDB UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL MCE MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 0.0570 | -59.5 | n/a | n/a | n/a | n/a | n/a | 5.5 | 30 |
GlaxoSmithKline | Assay Description Enzymatic activity was determined with fluorogenic substrate in the presence and absence of inhibitor. The fluorescence increase due to hydrolysis of... | Biochemistry 43: 14500-7 (2004) Article DOI: 10.1021/bi0488799 BindingDB Entry DOI: 10.7270/Q25M63WJ | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Delta-type opioid receptor (Rattus norvegicus (rat)) | BDBM21864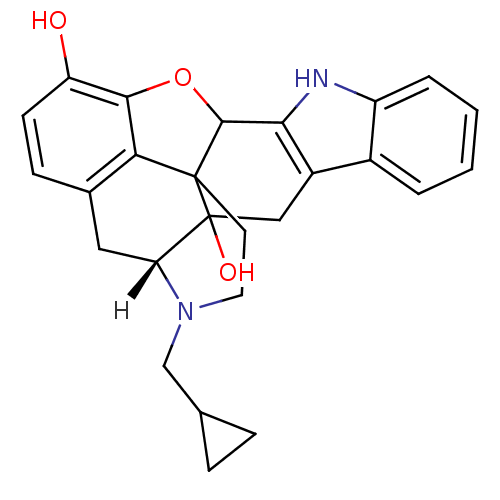 ((21R)-22-(cyclopropylmethyl)-14-oxa-11,22-diazahep...) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase PC cid PC sid UniChem Similars | Article PubMed | 0.240 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Southern Research Institute Curated by ChEMBL | Assay Description Displacement of [3H]DADLE from morphine-insensitive Opioid receptor delta 1 (presence of 50 nM morphine) of rat brain membranes | J Med Chem 41: 2872-81 (1998) Article DOI: 10.1021/jm980083i BindingDB Entry DOI: 10.7270/Q24F1RFN | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Delta-type opioid receptor (Rattus norvegicus (rat)) | BDBM21864 ((21R)-22-(cyclopropylmethyl)-14-oxa-11,22-diazahep...) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase PC cid PC sid UniChem Similars | Article PubMed | 0.290 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Southern Research Institute Curated by ChEMBL | Assay Description Displacement of [3H]DADLE from Opioid receptor delta 1 of rat brain membranes (DAMGO quenching mu receptor) | J Med Chem 41: 2872-81 (1998) Article DOI: 10.1021/jm980083i BindingDB Entry DOI: 10.7270/Q24F1RFN | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Delta-type opioid receptor (Rattus norvegicus (rat)) | BDBM21864 ((21R)-22-(cyclopropylmethyl)-14-oxa-11,22-diazahep...) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase PC cid PC sid UniChem Similars | Article PubMed | 0.410 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Southern Research Institute Curated by ChEMBL | Assay Description Binding affinity for Opioid receptor delta 1 was determined by inhibition of binding of [3H]DADLE (1.3-2.0 nM) to rat brain membranes | J Med Chem 42: 3527-38 (1999) Article DOI: 10.1021/jm990039i BindingDB Entry DOI: 10.7270/Q2QR4XSQ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Delta-type opioid receptor (Rattus norvegicus (rat)) | BDBM50065862 (22-cyclopropylmethyl-9-phenyl-14-oxa-11,22-diazahe...) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 0.460 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Southern Research Institute Curated by ChEMBL | Assay Description Displacement of [3H]DADLE from morphine-insensitive Opioid receptor delta 1 (presence of 50 nM morphine) of rat brain membranes | J Med Chem 41: 2872-81 (1998) Article DOI: 10.1021/jm980083i BindingDB Entry DOI: 10.7270/Q24F1RFN | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Delta-type opioid receptor (Rattus norvegicus (rat)) | BDBM50065854 (22-cyclopropylmethyl-9-phenoxy-14-oxa-11,22-diazah...) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 0.610 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Southern Research Institute Curated by ChEMBL | Assay Description Displacement of [3H]DADLE from morphine-insensitive Opioid receptor delta 1 (presence of 50 nM morphine) of rat brain membranes | J Med Chem 41: 2872-81 (1998) Article DOI: 10.1021/jm980083i BindingDB Entry DOI: 10.7270/Q24F1RFN | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Delta-type opioid receptor (Rattus norvegicus (rat)) | BDBM50065854 (22-cyclopropylmethyl-9-phenoxy-14-oxa-11,22-diazah...) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 0.710 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Southern Research Institute Curated by ChEMBL | Assay Description Displacement of [3H]DADLE from Opioid receptor delta 1 of rat brain membranes (DAMGO quenching mu receptor) | J Med Chem 41: 2872-81 (1998) Article DOI: 10.1021/jm980083i BindingDB Entry DOI: 10.7270/Q24F1RFN | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Delta-type opioid receptor (Rattus norvegicus (rat)) | BDBM50123273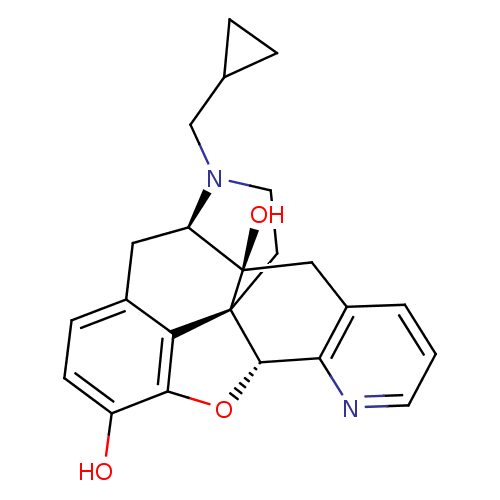 (19-cyclopropylmethyl-(2S,10R)-11-oxa-8,19-diazahex...) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 0.780 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Southern Research Institute Curated by ChEMBL | Assay Description Binding affinity for Opioid receptor delta 1 was determined by inhibition of binding of [3H]DADLE (1.3-2.0 nM) to rat brain membranes | J Med Chem 42: 3527-38 (1999) Article DOI: 10.1021/jm990039i BindingDB Entry DOI: 10.7270/Q2QR4XSQ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Delta-type opioid receptor (Rattus norvegicus (rat)) | BDBM50001196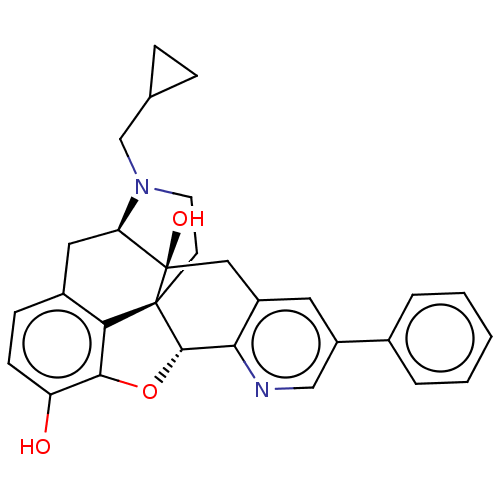 (CHEMBL610522) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 0.870 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Southern Research Institute Curated by ChEMBL | Assay Description Binding affinity for Opioid receptor delta 1 was determined by inhibition of binding of [3H]DADLE (1.3-2.0 nM) to rat brain membranes | J Med Chem 42: 3527-38 (1999) Article DOI: 10.1021/jm990039i BindingDB Entry DOI: 10.7270/Q2QR4XSQ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Mu-type opioid receptor (Rattus norvegicus (rat)) | BDBM50123273 (19-cyclopropylmethyl-(2S,10R)-11-oxa-8,19-diazahex...) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 1.5 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Southern Research Institute Curated by ChEMBL | Assay Description Binding affinity for Opioid receptor mu 1 was determined by inhibition of binding of [3H]DAMGO (1.4-3 nM) to rat brain membranes | J Med Chem 42: 3527-38 (1999) Article DOI: 10.1021/jm990039i BindingDB Entry DOI: 10.7270/Q2QR4XSQ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Delta-type opioid receptor (Rattus norvegicus (rat)) | BDBM50065857 (7-benzyloxy-22-cyclopropylmethyl-14-oxa-11,22-diaz...) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 1.70 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Southern Research Institute Curated by ChEMBL | Assay Description Displacement of [3H]DADLE from morphine-insensitive Opioid receptor delta 1 (presence of 50 nM morphine) of rat brain membranes | J Med Chem 41: 2872-81 (1998) Article DOI: 10.1021/jm980083i BindingDB Entry DOI: 10.7270/Q24F1RFN | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Delta-type opioid receptor (Rattus norvegicus (rat)) | BDBM50001190 (CHEMBL2112576) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 1.80 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Southern Research Institute Curated by ChEMBL | Assay Description Binding affinity for Opioid receptor delta 1 was determined by inhibition of binding of [3H]DADLE (1.3-2.0 nM) to rat brain membranes | J Med Chem 42: 3527-38 (1999) Article DOI: 10.1021/jm990039i BindingDB Entry DOI: 10.7270/Q2QR4XSQ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Gonadotropin-releasing hormone receptor (Homo sapiens (Human)) | BDBM50152714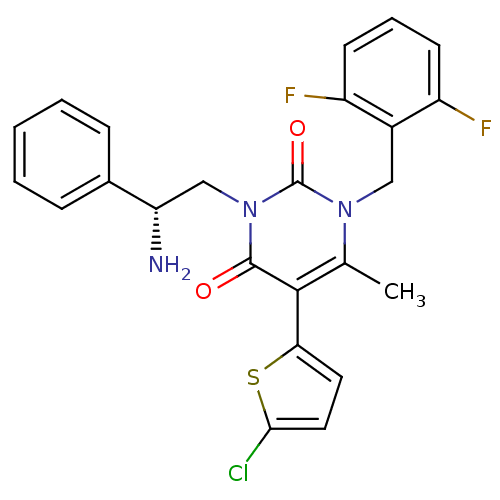 ((R)-1-(2,6-difluorobenzyl)-3-(2-amino-2-phenylethy...) | PDB KEGG UniProtKB/SwissProt DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 2 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Neurocrine Biosciences Inc. Curated by ChEMBL | Assay Description Binding affinity towards cloned human Gonadotropin-releasing hormone receptor expressed in RBL cells was determined by using [125I]-GnRH as radioliga... | Bioorg Med Chem Lett 14: 4967-73 (2004) Article DOI: 10.1016/j.bmcl.2004.07.022 BindingDB Entry DOI: 10.7270/Q2FF3RT4 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Delta-type opioid receptor (Rattus norvegicus (rat)) | BDBM50065862 (22-cyclopropylmethyl-9-phenyl-14-oxa-11,22-diazahe...) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 2.10 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Southern Research Institute Curated by ChEMBL | Assay Description Displacement of [3H]DADLE from Opioid receptor delta 1 of rat brain membranes (DAMGO quenching mu receptor) | J Med Chem 41: 2872-81 (1998) Article DOI: 10.1021/jm980083i BindingDB Entry DOI: 10.7270/Q24F1RFN | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Delta-type opioid receptor (Rattus norvegicus (rat)) | BDBM50001195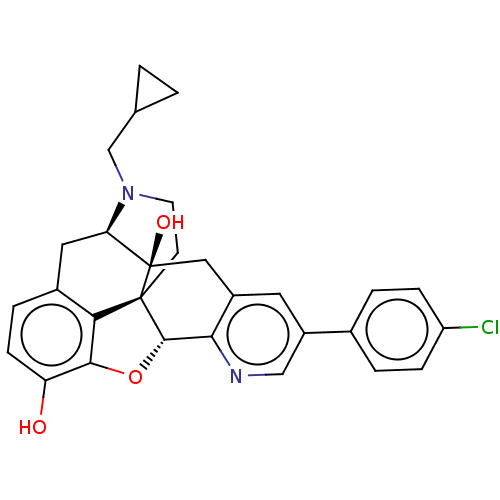 (CHEMBL610261) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 2.20 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Southern Research Institute Curated by ChEMBL | Assay Description Binding affinity for Opioid receptor delta 1 was determined by inhibition of binding of [3H]DADLE (1.3-2.0 nM) to rat brain membranes | J Med Chem 42: 3527-38 (1999) Article DOI: 10.1021/jm990039i BindingDB Entry DOI: 10.7270/Q2QR4XSQ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [489-587,M535I,L552P,A560V,V571F,I573V] (Human immunodeficiency virus type 1) | BDBM577 ((3S)-oxolan-3-yl N-[(2S,3R)-4-[(4-aminobenzene)(2-...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL MCE MMDB PC cid PC sid PDB UniChem Patents Similars | Article PubMed | 2.30 | -50.1 | n/a | n/a | n/a | n/a | n/a | 5.5 | 30 |
GlaxoSmithKline | Assay Description Enzymatic activity was determined with fluorogenic substrate in the presence and absence of inhibitor. The fluorescence increase due to hydrolysis of... | Biochemistry 43: 14500-7 (2004) Article DOI: 10.1021/bi0488799 BindingDB Entry DOI: 10.7270/Q25M63WJ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Mu-type opioid receptor (Rattus norvegicus (rat)) | BDBM60212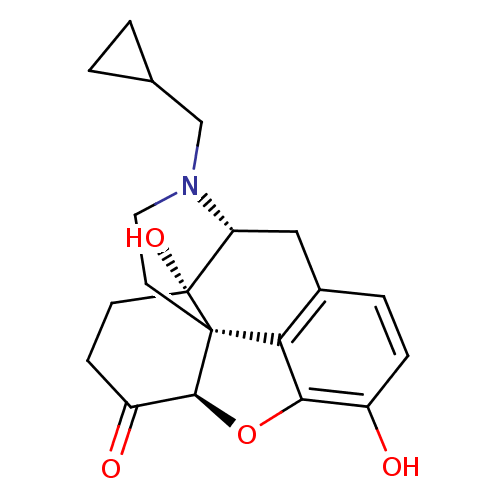 ((4R,4aS,7aR,12bS)-3-(cyclopropylmethyl)-4a,9-bis(o...) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase DrugBank MCE KEGG PC cid PC sid UniChem Similars | Article PubMed | 2.5 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Southern Research Institute Curated by ChEMBL | Assay Description Binding affinity for Opioid receptor mu 1 was determined by inhibition of binding of [3H]DAMGO (1.4-3 nM) to rat brain membranes | J Med Chem 42: 3527-38 (1999) Article DOI: 10.1021/jm990039i BindingDB Entry DOI: 10.7270/Q2QR4XSQ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Delta-type opioid receptor (Rattus norvegicus (rat)) | BDBM50065858 (9-benzyloxy-22-cyclopropylmethyl-14-oxa-11,22-diaz...) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 2.5 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Southern Research Institute Curated by ChEMBL | Assay Description Displacement of [3H]DADLE from morphine-insensitive Opioid receptor delta 1 (presence of 50 nM morphine) of rat brain membranes | J Med Chem 41: 2872-81 (1998) Article DOI: 10.1021/jm980083i BindingDB Entry DOI: 10.7270/Q24F1RFN | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Gonadotropin-releasing hormone receptor (Homo sapiens (Human)) | BDBM50152708 (3-((R)-2-Amino-2-phenyl-ethyl)-1-(2,6-difluoro-ben...) | PDB KEGG UniProtKB/SwissProt DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Neurocrine Biosciences Inc. Curated by ChEMBL | Assay Description Binding affinity towards cloned human Gonadotropin-releasing hormone receptor expressed in RBL cells was determined by using [125I]-GnRH as radioliga... | Bioorg Med Chem Lett 14: 4967-73 (2004) Article DOI: 10.1016/j.bmcl.2004.07.022 BindingDB Entry DOI: 10.7270/Q2FF3RT4 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Delta-type opioid receptor (Rattus norvegicus (rat)) | BDBM50065858 (9-benzyloxy-22-cyclopropylmethyl-14-oxa-11,22-diaz...) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 3.10 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Southern Research Institute Curated by ChEMBL | Assay Description Displacement of [3H]DADLE from Opioid receptor delta 1 of rat brain membranes (DAMGO quenching mu receptor) | J Med Chem 41: 2872-81 (1998) Article DOI: 10.1021/jm980083i BindingDB Entry DOI: 10.7270/Q24F1RFN | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Delta-type opioid receptor (MOUSE) | BDBM50001196 (CHEMBL610522) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 3.40 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Southern Research Institute Curated by ChEMBL | Assay Description Antagonist potency measured as Ke = [antagonist]/(IC50 ratio-1) against delta opioid receptor from mouse vas deferens (MVD) using DPDPE | J Med Chem 42: 3527-38 (1999) Article DOI: 10.1021/jm990039i BindingDB Entry DOI: 10.7270/Q2QR4XSQ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Delta-type opioid receptor (Rattus norvegicus (rat)) | BDBM50001173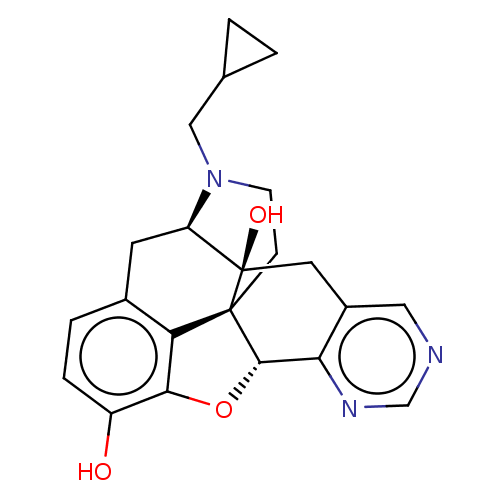 (CHEMBL2112572) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 3.5 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Southern Research Institute Curated by ChEMBL | Assay Description Binding affinity for Opioid receptor delta 1 was determined by inhibition of binding of [3H]DADLE (1.3-2.0 nM) to rat brain membranes | J Med Chem 42: 3527-38 (1999) Article DOI: 10.1021/jm990039i BindingDB Entry DOI: 10.7270/Q2QR4XSQ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Delta-type opioid receptor (Rattus norvegicus (rat)) | BDBM50065863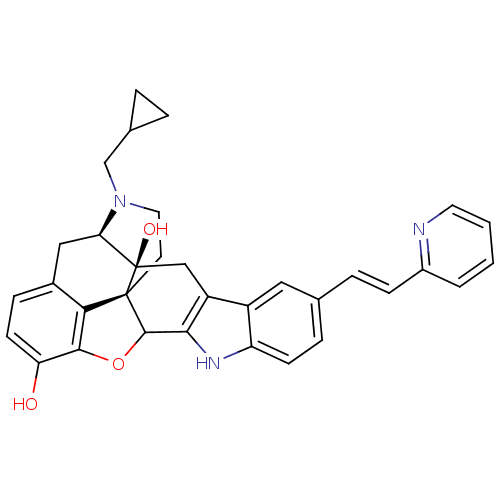 (22-cyclopropylmethyl-7-[2-(2-pyridyl)-(E)-1-etheny...) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 3.70 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Southern Research Institute Curated by ChEMBL | Assay Description Displacement of [3H]DADLE from Opioid receptor delta 1 of rat brain membranes (DAMGO quenching mu receptor) | J Med Chem 41: 2872-81 (1998) Article DOI: 10.1021/jm980083i BindingDB Entry DOI: 10.7270/Q24F1RFN | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Delta-type opioid receptor (Rattus norvegicus (rat)) | BDBM50065863 (22-cyclopropylmethyl-7-[2-(2-pyridyl)-(E)-1-etheny...) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Southern Research Institute Curated by ChEMBL | Assay Description Displacement of [3H]DADLE from morphine-insensitive Opioid receptor delta 1 (presence of 50 nM morphine) of rat brain membranes | J Med Chem 41: 2872-81 (1998) Article DOI: 10.1021/jm980083i BindingDB Entry DOI: 10.7270/Q24F1RFN | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Mu-type opioid receptor (Rattus norvegicus (rat)) | BDBM50001173 (CHEMBL2112572) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 4.20 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Southern Research Institute Curated by ChEMBL | Assay Description Binding affinity for Opioid receptor mu 1 was determined by inhibition of binding of [3H]DAMGO (1.4-3 nM) to rat brain membranes | J Med Chem 42: 3527-38 (1999) Article DOI: 10.1021/jm990039i BindingDB Entry DOI: 10.7270/Q2QR4XSQ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Delta-type opioid receptor (Rattus norvegicus (rat)) | BDBM50065857 (7-benzyloxy-22-cyclopropylmethyl-14-oxa-11,22-diaz...) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 4.40 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Southern Research Institute Curated by ChEMBL | Assay Description Displacement of [3H]DADLE from Opioid receptor delta 1 of rat brain membranes (DAMGO quenching mu receptor) | J Med Chem 41: 2872-81 (1998) Article DOI: 10.1021/jm980083i BindingDB Entry DOI: 10.7270/Q24F1RFN | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Delta-type opioid receptor (Rattus norvegicus (rat)) | BDBM50065856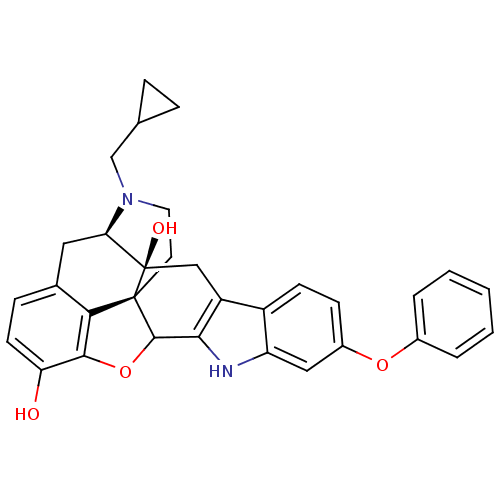 (22-cyclopropylmethyl-8-phenoxy-14-oxa-11,22-diazah...) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 4.90 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Southern Research Institute Curated by ChEMBL | Assay Description Displacement of [3H]DADLE from morphine-insensitive Opioid receptor delta 1 (presence of 50 nM morphine) of rat brain membranes | J Med Chem 41: 2872-81 (1998) Article DOI: 10.1021/jm980083i BindingDB Entry DOI: 10.7270/Q24F1RFN | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Delta-type opioid receptor (Rattus norvegicus (rat)) | BDBM50065864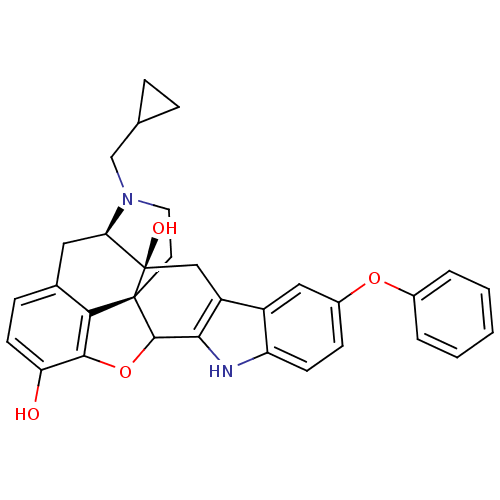 (22-cyclopropylmethyl-7-phenoxy-14-oxa-11,22-diazah...) | PDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 4.90 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Southern Research Institute Curated by ChEMBL | Assay Description Displacement of [3H]DADLE from morphine-insensitive Opioid receptor delta 1 (presence of 50 nM morphine) of rat brain membranes | J Med Chem 41: 2872-81 (1998) Article DOI: 10.1021/jm980083i BindingDB Entry DOI: 10.7270/Q24F1RFN | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Dimer of Gag-Pol polyprotein [489-587,I539V] (Human immunodeficiency virus type 1) | BDBM577 ((3S)-oxolan-3-yl N-[(2S,3R)-4-[(4-aminobenzene)(2-...) | PDB MMDB UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL MCE MMDB PC cid PC sid PDB UniChem Patents Similars | MMDB PDB Article PubMed | 4.90 | -48.2 | n/a | n/a | n/a | n/a | n/a | 5.5 | 30 |
GlaxoSmithKline | Assay Description Enzymatic activity was determined with fluorogenic substrate in the presence and absence of inhibitor. The fluorescence increase due to hydrolysis of... | Biochemistry 43: 14500-7 (2004) Article DOI: 10.1021/bi0488799 BindingDB Entry DOI: 10.7270/Q25M63WJ | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Gonadotropin-releasing hormone receptor (Homo sapiens (Human)) | BDBM50132726 ((R)-1-(2,6-difluorobenzyl)-5-(3-methoxyphenyl)-6-m...) | PDB KEGG UniProtKB/SwissProt DrugBank antibodypedia GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | 5 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
Neurocrine Biosciences Inc. Curated by ChEMBL | Assay Description Binding affinity towards cloned human Gonadotropin-releasing hormone receptor expressed in RBL cells was determined by using [125I]-GnRH as radioliga... | Bioorg Med Chem Lett 14: 4967-73 (2004) Article DOI: 10.1016/j.bmcl.2004.07.022 BindingDB Entry DOI: 10.7270/Q2FF3RT4 | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Displayed 1 to 50 (of 885 total ) | Next | Last >> |