

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330319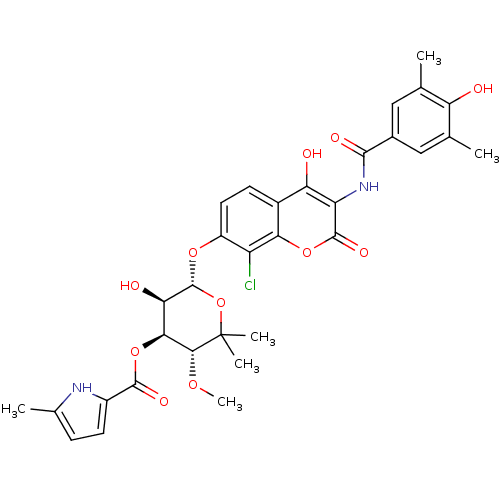 ((3R,4S,5R,6S)-6-(8-chloro-4-hydroxy-3-(4-hydroxy-3...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 17 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as DNA triple helix formation by supercoiling assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330315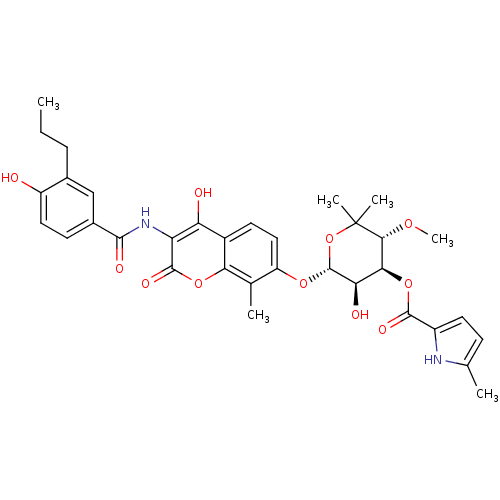 ((3R,4S,5R,6R)-5-hydroxy-6-(4-hydroxy-3-(4-hydroxy-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 19 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as DNA triple helix formation by supercoiling assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330314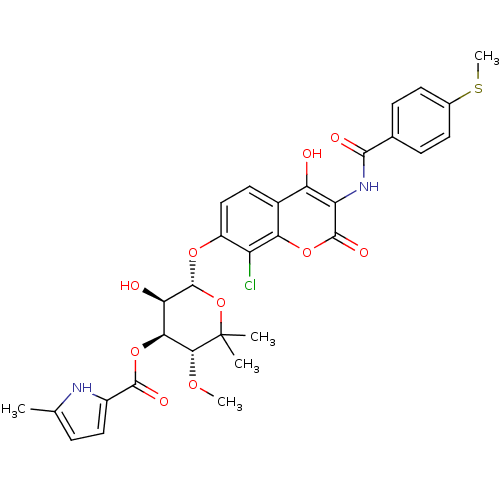 ((3R,4S,5R,6S)-6-(8-chloro-4-hydroxy-3-(4-(methylth...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 20 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as DNA triple helix formation by supercoiling assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330309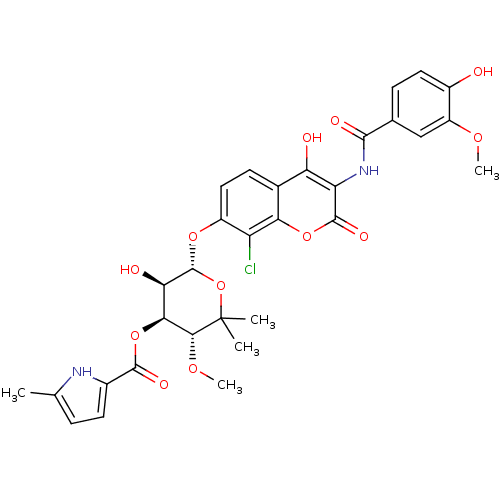 ((3R,4S,5R,6S)-6-(8-chloro-4-hydroxy-3-(4-hydroxy-3...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 22 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as DNA triple helix formation by supercoiling assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330313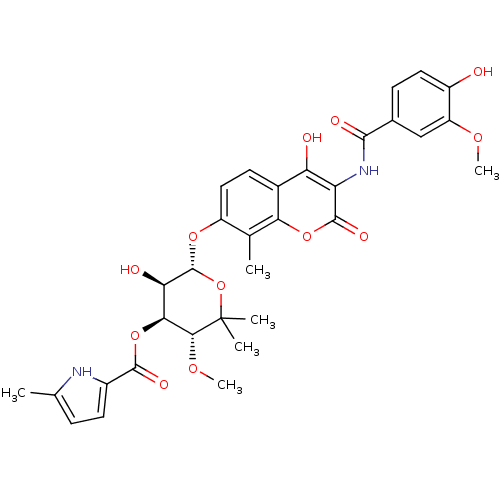 ((3R,4S,5R,6R)-5-hydroxy-6-(4-hydroxy-3-(4-hydroxy-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 25 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as DNA triple helix formation by supercoiling assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330307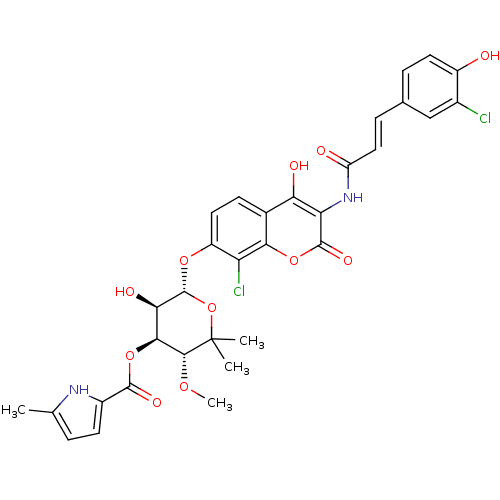 ((3R,4S,5R,6S)-6-(8-chloro-3-((E)-3-(3-chloro-4-hyd...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 32 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as DNA triple helix formation by supercoiling assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330312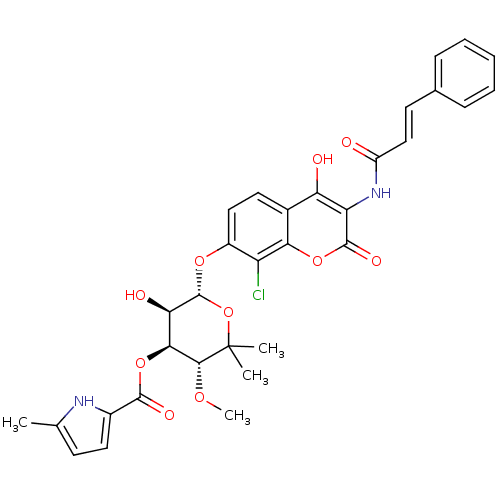 ((3R,4S,5R,6S)-6-(8-chloro-3-cinnamamido-4-hydroxy-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 34 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as DNA triple helix formation by supercoiling assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330310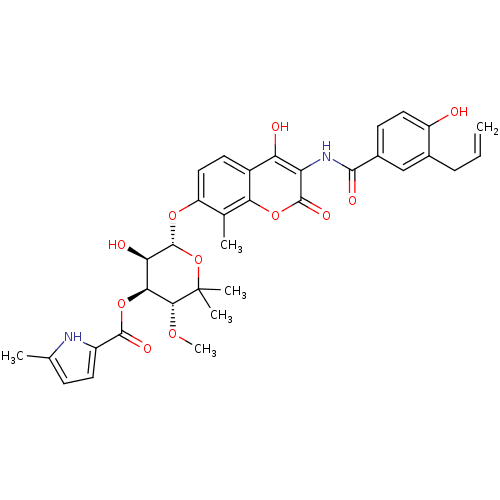 ((3R,4S,5R,6R)-6-(3-(3-allyl-4-hydroxybenzamido)-4-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 34 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as DNA triple helix formation by supercoiling assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330316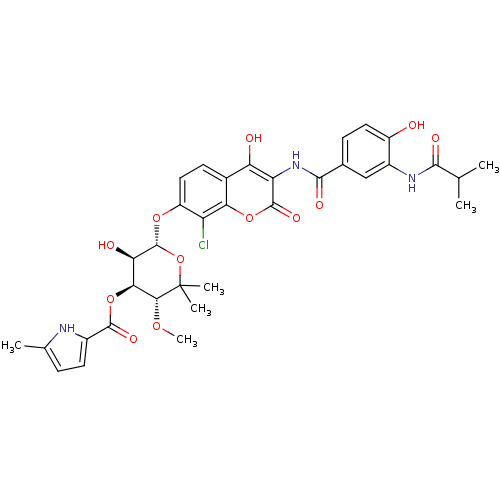 ((3R,4S,5R,6S)-6-(8-chloro-4-hydroxy-3-(4-hydroxy-3...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 41 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as DNA triple helix formation by supercoiling assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330302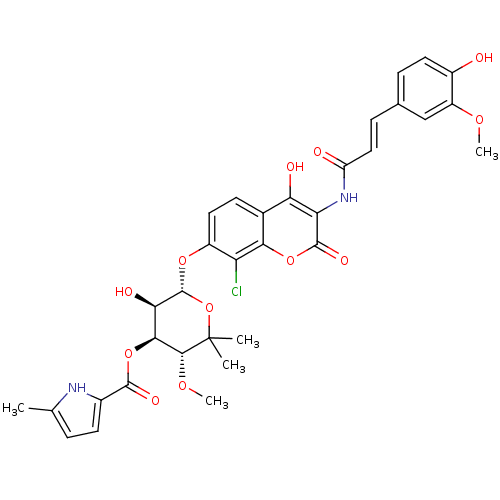 ((3R,4S,5R,6S)-6-(8-chloro-4-hydroxy-3-((E)-3-(4-hy...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 43 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as DNA triple helix formation by supercoiling assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330306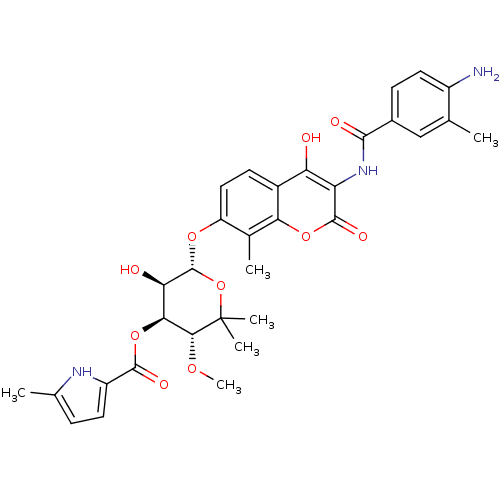 ((3R,4S,5R,6R)-6-(3-(4-amino-3-methylbenzamido)-4-h...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 44 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as DNA triple helix formation by supercoiling assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330304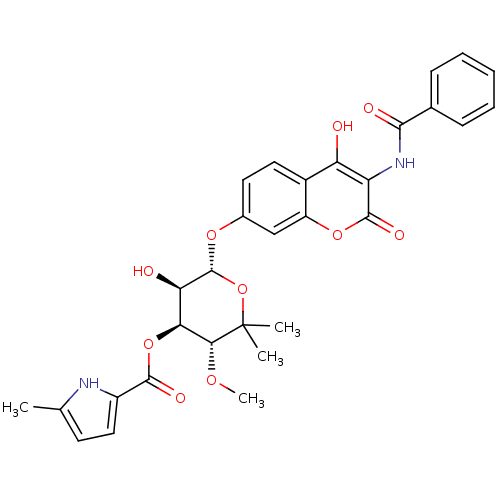 ((3R,4S,5R,6R)-6-(3-benzamido-4-hydroxy-2-oxo-2H-ch...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 50 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as DNA triple helix formation by supercoiling assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330319 ((3R,4S,5R,6S)-6-(8-chloro-4-hydroxy-3-(4-hydroxy-3...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 65 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as ATP hydrolysis by ATPase assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330318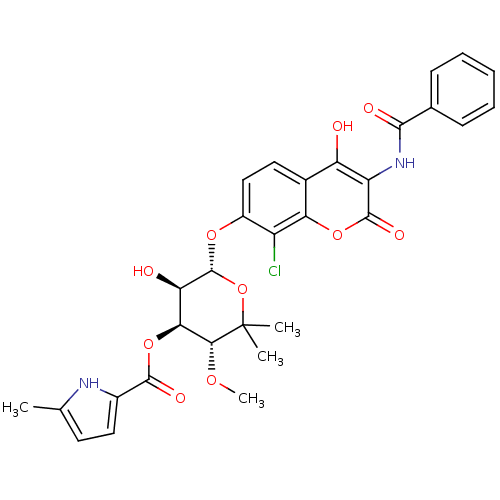 ((3R,4S,5R,6S)-6-(3-benzamido-8-chloro-4-hydroxy-2-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 72 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as ATP hydrolysis by ATPase assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330316 ((3R,4S,5R,6S)-6-(8-chloro-4-hydroxy-3-(4-hydroxy-3...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 73 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as ATP hydrolysis by ATPase assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330317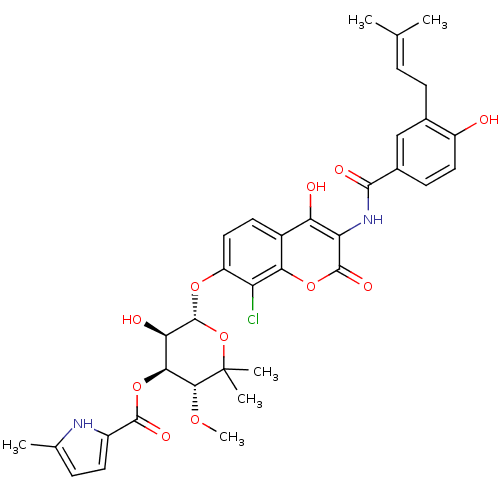 (5-Methyl-1H-pyrrole-2-carboxylic acid (3R,4S,5R,6S...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL MCE KEGG MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | n/a | n/a | 73 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as ATP hydrolysis by ATPase assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330305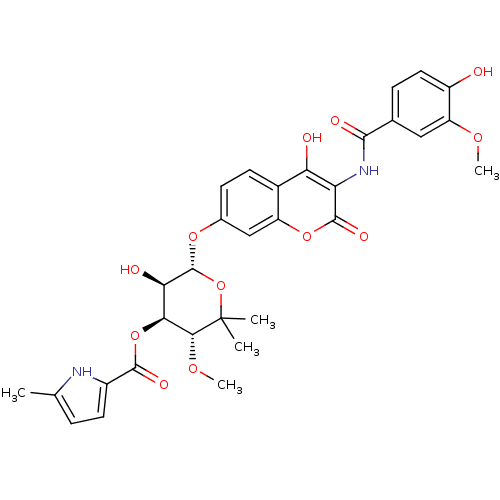 ((3R,4S,5R,6R)-5-hydroxy-6-(4-hydroxy-3-(4-hydroxy-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 75 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as DNA triple helix formation by supercoiling assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330315 ((3R,4S,5R,6R)-5-hydroxy-6-(4-hydroxy-3-(4-hydroxy-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 84 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as ATP hydrolysis by ATPase assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330314 ((3R,4S,5R,6S)-6-(8-chloro-4-hydroxy-3-(4-(methylth...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 84 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as ATP hydrolysis by ATPase assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330313 ((3R,4S,5R,6R)-5-hydroxy-6-(4-hydroxy-3-(4-hydroxy-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 86 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as ATP hydrolysis by ATPase assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330312 ((3R,4S,5R,6S)-6-(8-chloro-3-cinnamamido-4-hydroxy-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 90 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as ATP hydrolysis by ATPase assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330311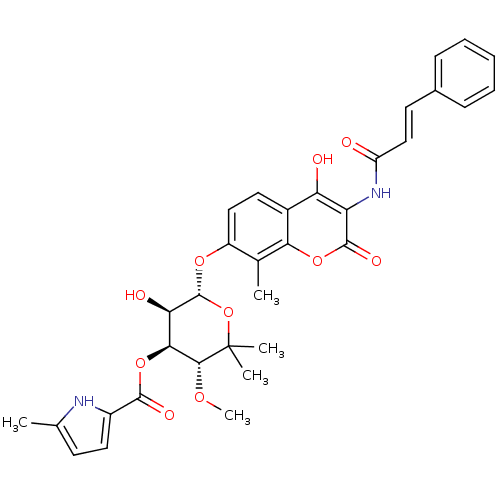 ((3R,4S,5R,6R)-6-(3-cinnamamido-4-hydroxy-8-methyl-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 106 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as ATP hydrolysis by ATPase assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330299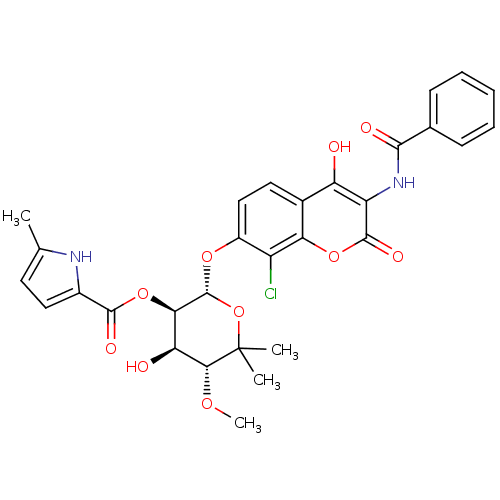 ((2S,3R,4R,5R)-2-(3-benzamido-8-chloro-4-hydroxy-2-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 110 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as DNA triple helix formation by supercoiling assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330310 ((3R,4S,5R,6R)-6-(3-(3-allyl-4-hydroxybenzamido)-4-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 116 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as ATP hydrolysis by ATPase assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330303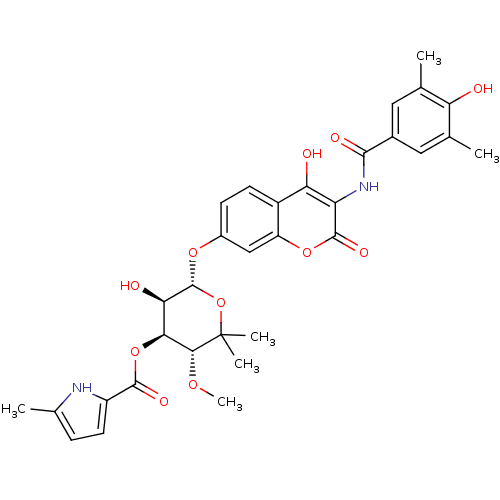 ((3R,4S,5R,6R)-5-hydroxy-6-(4-hydroxy-3-(4-hydroxy-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 120 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as DNA triple helix formation by supercoiling assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330309 ((3R,4S,5R,6S)-6-(8-chloro-4-hydroxy-3-(4-hydroxy-3...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 122 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as ATP hydrolysis by ATPase assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330308 ((3R,4S,5R,6R)-6-(3-benzamido-4-hydroxy-8-methyl-2-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 131 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as ATP hydrolysis by ATPase assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330307 ((3R,4S,5R,6S)-6-(8-chloro-3-((E)-3-(3-chloro-4-hyd...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 137 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as ATP hydrolysis by ATPase assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330295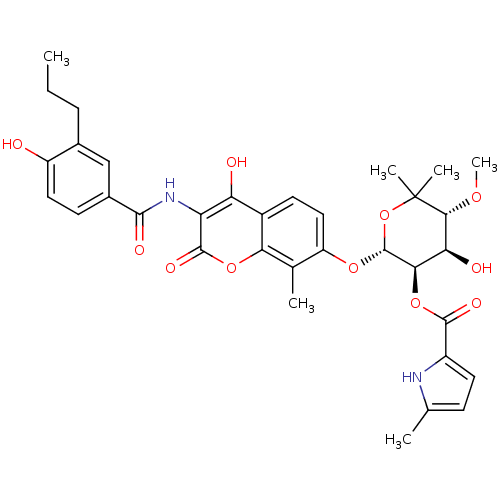 ((2R,3R,4R,5R)-4-hydroxy-2-(4-hydroxy-3-(4-hydroxy-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 140 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as DNA triple helix formation by supercoiling assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330306 ((3R,4S,5R,6R)-6-(3-(4-amino-3-methylbenzamido)-4-h...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 152 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as ATP hydrolysis by ATPase assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50423645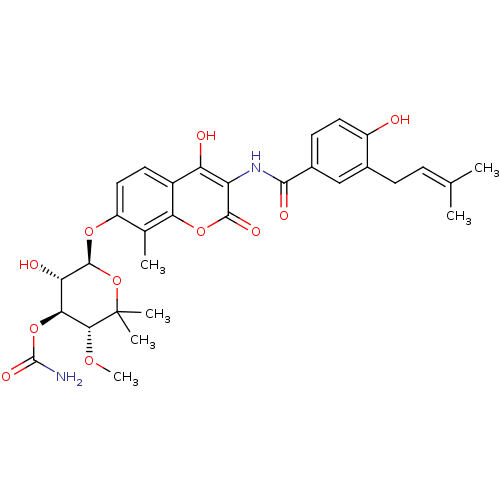 (Albamycin | Cathomycin | NOVOBIOCIN) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 154 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as ATP hydrolysis by ATPase assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330305 ((3R,4S,5R,6R)-5-hydroxy-6-(4-hydroxy-3-(4-hydroxy-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 185 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as ATP hydrolysis by ATPase assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330294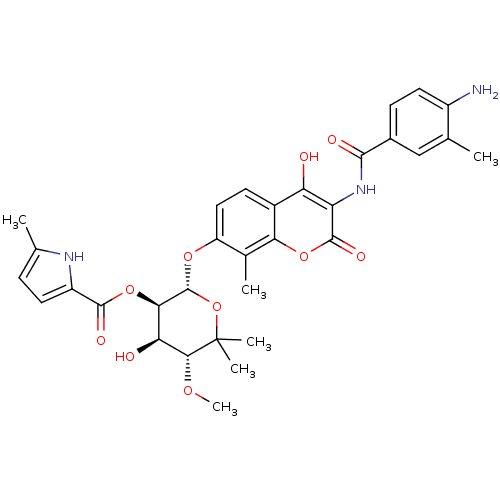 ((2R,3R,4R,5R)-2-(3-(4-amino-3-methylbenzamido)-4-h...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 190 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as DNA triple helix formation by supercoiling assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50423645 (Albamycin | Cathomycin | NOVOBIOCIN) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 200 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as DNA triple helix formation by supercoiling assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330304 ((3R,4S,5R,6R)-6-(3-benzamido-4-hydroxy-2-oxo-2H-ch...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 219 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as ATP hydrolysis by ATPase assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330303 ((3R,4S,5R,6R)-5-hydroxy-6-(4-hydroxy-3-(4-hydroxy-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 229 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as ATP hydrolysis by ATPase assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330302 ((3R,4S,5R,6S)-6-(8-chloro-4-hydroxy-3-((E)-3-(4-hy...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 263 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as ATP hydrolysis by ATPase assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330301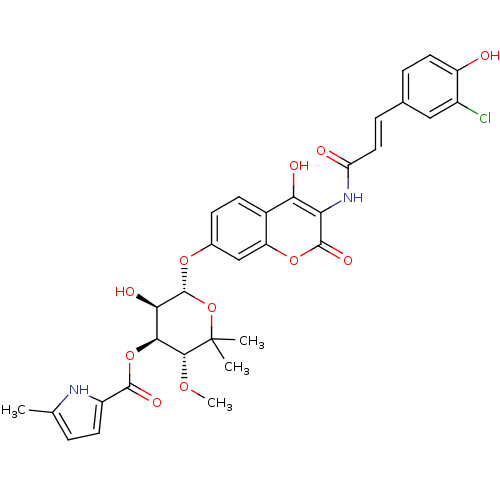 ((3R,4S,5R,6R)-6-(3-((E)-3-(3-chloro-4-hydroxypheny...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 276 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as ATP hydrolysis by ATPase assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330301 ((3R,4S,5R,6R)-6-(3-((E)-3-(3-chloro-4-hydroxypheny...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 280 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as DNA triple helix formation by supercoiling assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330300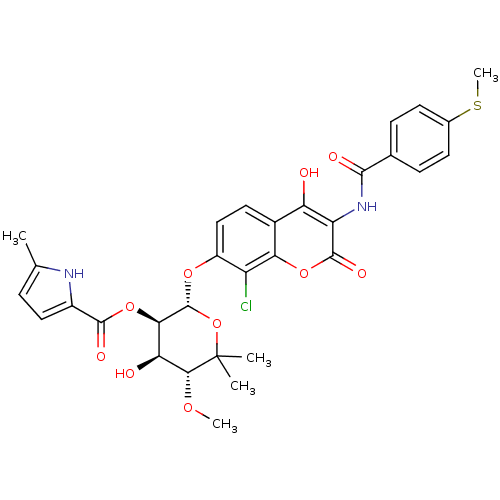 ((2S,3R,4R,5R)-2-(8-chloro-4-hydroxy-3-(4-(methylth...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 280 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as ATP hydrolysis by ATPase assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330299 ((2S,3R,4R,5R)-2-(3-benzamido-8-chloro-4-hydroxy-2-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 297 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as ATP hydrolysis by ATPase assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330298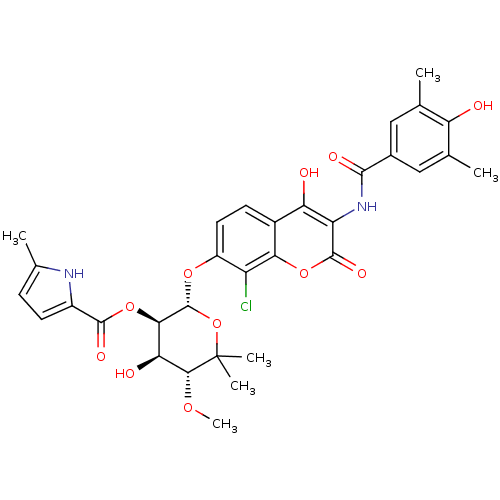 ((2S,3R,4R,5R)-2-(8-chloro-4-hydroxy-3-(4-hydroxy-3...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 320 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as DNA triple helix formation by supercoiling assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330298 ((2S,3R,4R,5R)-2-(8-chloro-4-hydroxy-3-(4-hydroxy-3...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 376 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as ATP hydrolysis by ATPase assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330293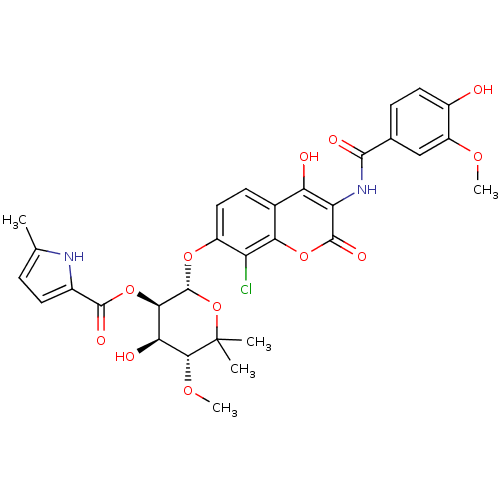 ((2S,3R,4R,5R)-2-(8-chloro-4-hydroxy-3-(4-hydroxy-3...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 460 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as DNA triple helix formation by supercoiling assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330292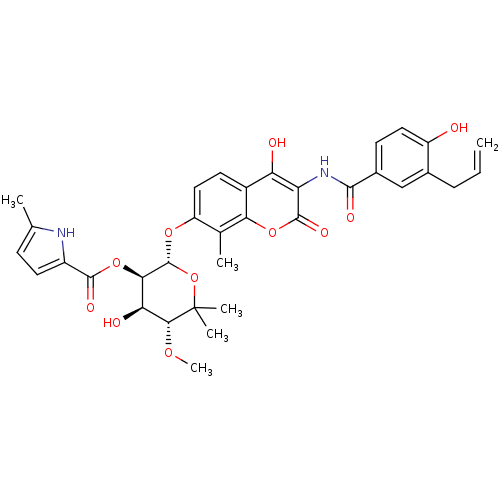 ((2R,3R,4R,5R)-2-(3-(3-allyl-4-hydroxybenzamido)-4-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 460 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as DNA triple helix formation by supercoiling assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330297 ((2S,3R,4R,5R)-2-(8-chloro-3-cinnamamido-4-hydroxy-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 598 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as ATP hydrolysis by ATPase assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330295 ((2R,3R,4R,5R)-4-hydroxy-2-(4-hydroxy-3-(4-hydroxy-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 758 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as ATP hydrolysis by ATPase assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330294 ((2R,3R,4R,5R)-2-(3-(4-amino-3-methylbenzamido)-4-h...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 784 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as ATP hydrolysis by ATPase assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330293 ((2S,3R,4R,5R)-2-(8-chloro-4-hydroxy-3-(4-hydroxy-3...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 891 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as ATP hydrolysis by ATPase assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| DNA gyrase subunit B (Escherichia coli (strain K12)) | BDBM50330297 ((2S,3R,4R,5R)-2-(8-chloro-3-cinnamamido-4-hydroxy-...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem Similars | Article PubMed | n/a | n/a | 1.20E+3 | n/a | n/a | n/a | n/a | n/a | n/a |
Universität Tübingen Curated by ChEMBL | Assay Description Inhibition of Escherichia coli JM109 DNA gyrase subunit B assessed as DNA triple helix formation by supercoiling assay | Antimicrob Agents Chemother 52: 1982-90 (2008) Article DOI: 10.1128/AAC.01235-07 BindingDB Entry DOI: 10.7270/Q2PG1S0J | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Displayed 1 to 50 (of 80 total ) | Next | Last >> |