

 Search and Browse
Search and Browse
 Download
Download
 Enter Data
Enter Data
| Target/Host (Institution) | Ligand | Target/Host Links | Ligand Links | Trg + Lig Links | Ki nM | ΔG° kJ/mole | IC50 nM | Kd nM | EC50/IC50 nM | koff s-1 | kon M-1s-1 | pH | Temp °C |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Androgen receptor (Rattus norvegicus (Rat)) | BDBM18161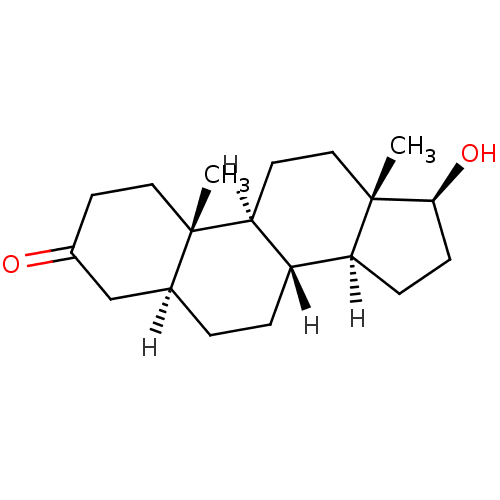 ((1S,2S,7S,10R,11S,14S,15S)-14-hydroxy-2,15-dimethy...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase CHEMBL KEGG MMDB PC cid PC sid PDB UniChem Patents Similars | PDB Article PubMed | 0.270 | -50.8 | n/a | n/a | n/a | n/a | n/a | 7.4 | 4 |
The Ohio State University | Assay Description The Ki values were determined by the application of the Cheng-Prusoff equation: Ki = (IC50 x Kd)/(Kd+[L]) where [L] is the concentration of [3H]MIB (... | Bioorg Med Chem Lett 18: 5567-70 (2008) Article DOI: 10.1016/j.bmcl.2008.09.002 BindingDB Entry DOI: 10.7270/Q2W0948B | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Androgen receptor (Rattus norvegicus (Rat)) | BDBM26260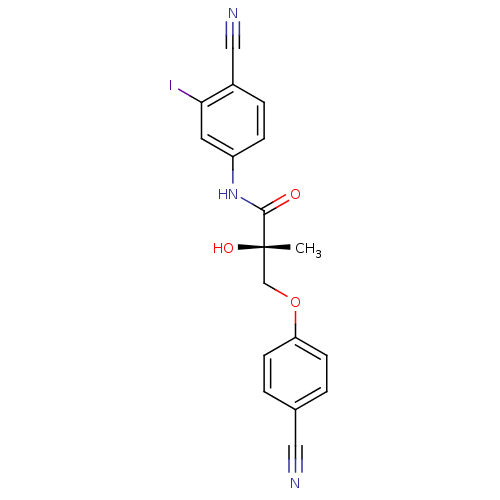 ((2S)-N-(4-cyano-3-iodophenyl)-3-(4-cyanophenoxy)-2...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | DrugBank MMDB PC cid PC sid PDB UniChem | MMDB PDB Article PubMed | 0.540 | -49.2 | n/a | n/a | n/a | n/a | n/a | 7.4 | 4 |
The Ohio State University | Assay Description The Ki values were determined by the application of the Cheng-Prusoff equation: Ki = (IC50 x Kd)/(Kd+[L]) where [L] is the concentration of [3H]MIB (... | Bioorg Med Chem Lett 18: 5567-70 (2008) Article DOI: 10.1016/j.bmcl.2008.09.002 BindingDB Entry DOI: 10.7270/Q2W0948B | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Androgen receptor (Rattus norvegicus (Rat)) | BDBM26262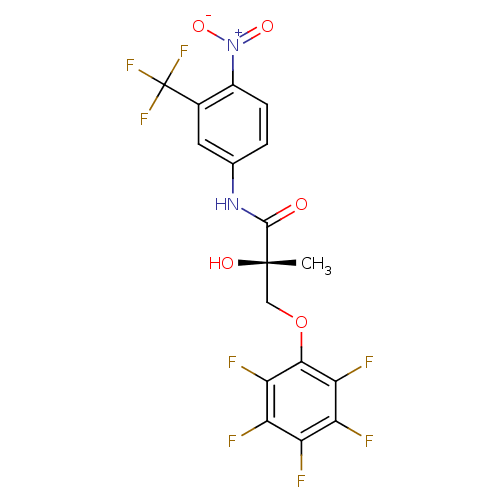 ((2S)-2-hydroxy-2-methyl-N-[4-nitro-3-(trifluoromet...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | DrugBank MMDB PC cid PC sid PDB UniChem Patents Similars | MMDB PDB Article PubMed | 1.40 | -47.0 | n/a | n/a | n/a | n/a | n/a | 7.4 | 4 |
The Ohio State University | Assay Description The Ki values were determined by the application of the Cheng-Prusoff equation: Ki = (IC50 x Kd)/(Kd+[L]) where [L] is the concentration of [3H]MIB (... | Bioorg Med Chem Lett 18: 5567-70 (2008) Article DOI: 10.1016/j.bmcl.2008.09.002 BindingDB Entry DOI: 10.7270/Q2W0948B | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Androgen receptor (Rattus norvegicus (Rat)) | BDBM26261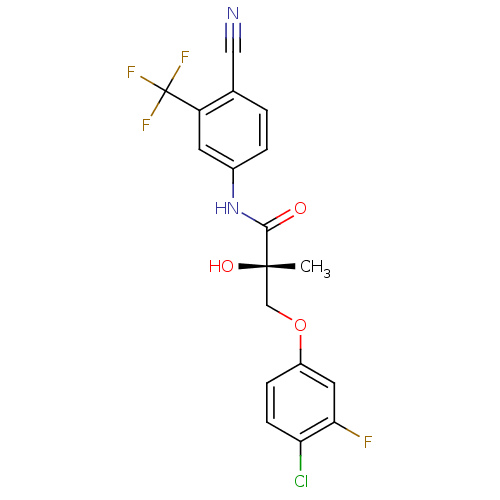 ((2S)-3-(4-chloro-3-fluorophenoxy)-N-[4-cyano-3-(tr...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | 1.70 | -46.5 | n/a | n/a | n/a | n/a | n/a | 7.4 | 4 |
The Ohio State University | Assay Description The Ki values were determined by the application of the Cheng-Prusoff equation: Ki = (IC50 x Kd)/(Kd+[L]) where [L] is the concentration of [3H]MIB (... | Bioorg Med Chem Lett 18: 5567-70 (2008) Article DOI: 10.1016/j.bmcl.2008.09.002 BindingDB Entry DOI: 10.7270/Q2W0948B | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Androgen receptor (Rattus norvegicus (Rat)) | BDBM26259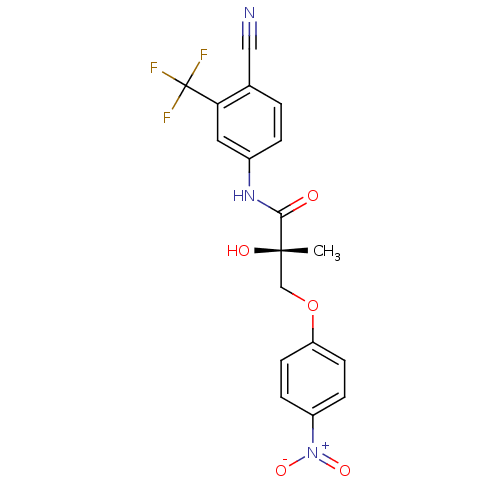 ((2S)-N-[4-cyano-3-(trifluoromethyl)phenyl]-2-hydro...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | MMDB PC cid PC sid UniChem Similars | MMDB Article PubMed | 2.5 | -45.6 | n/a | n/a | n/a | n/a | n/a | 7.4 | 4 |
The Ohio State University | Assay Description The Ki values were determined by the application of the Cheng-Prusoff equation: Ki = (IC50 x Kd)/(Kd+[L]) where [L] is the concentration of [3H]MIB (... | Bioorg Med Chem Lett 18: 5567-70 (2008) Article DOI: 10.1016/j.bmcl.2008.09.002 BindingDB Entry DOI: 10.7270/Q2W0948B | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| Androgen receptor (Rattus norvegicus (Rat)) | BDBM18665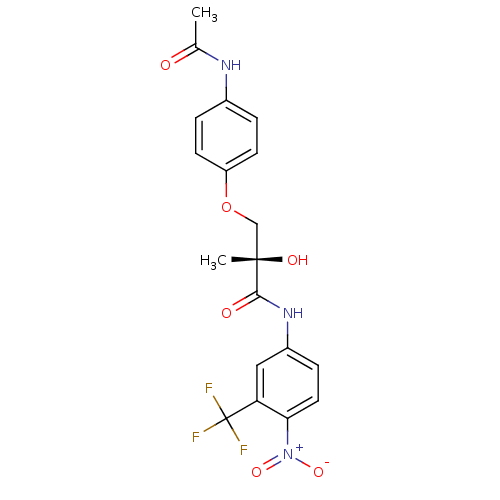 ((2S)-3-(4-acetamidophenoxy)-2-hydroxy-2-methyl-N-[...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE MMDB PC cid PC sid PDB UniChem Patents Similars | MMDB PDB Article PubMed | 4 | -44.6 | n/a | n/a | n/a | n/a | n/a | 7.4 | 4 |
The Ohio State University | Assay Description The Ki values were determined by the application of the Cheng-Prusoff equation: Ki = (IC50 x Kd)/(Kd+[L]) where [L] is the concentration of [3H]MIB (... | Bioorg Med Chem Lett 18: 5567-70 (2008) Article DOI: 10.1016/j.bmcl.2008.09.002 BindingDB Entry DOI: 10.7270/Q2W0948B | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Androgen receptor (Rattus norvegicus (Rat)) | BDBM18663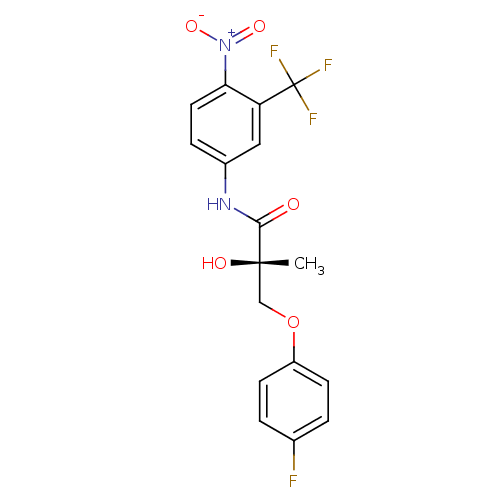 ((2S)-3-(4-fluorophenoxy)-2-hydroxy-2-methyl-N-[4-n...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | CHEMBL DrugBank MMDB PC cid PC sid PDB UniChem Patents Similars | MMDB PDB Article PubMed | 6.10 | -43.6 | n/a | n/a | n/a | n/a | n/a | 7.4 | 4 |
The Ohio State University | Assay Description The Ki values were determined by the application of the Cheng-Prusoff equation: Ki = (IC50 x Kd)/(Kd+[L]) where [L] is the concentration of [3H]MIB (... | Bioorg Med Chem Lett 18: 5567-70 (2008) Article DOI: 10.1016/j.bmcl.2008.09.002 BindingDB Entry DOI: 10.7270/Q2W0948B | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| Androgen receptor (Rattus norvegicus (Rat)) | BDBM18678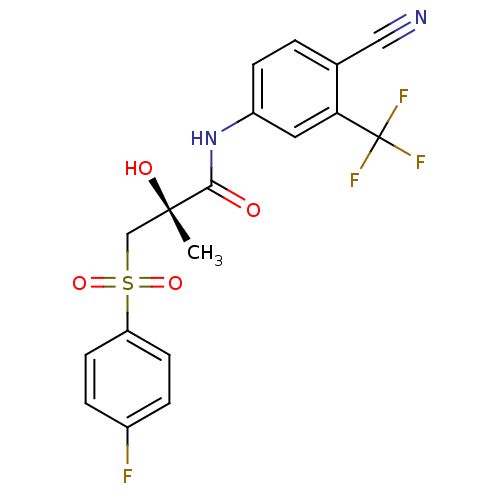 ((2R)-N-[4-cyano-3-(trifluoromethyl)phenyl]-3-[(4-f...) | PDB MMDB Reactome pathway KEGG UniProtKB/SwissProt B.MOAD GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE KEGG MMDB PC cid PC sid PDB UniChem Patents Similars | MMDB PDB Article PubMed | 11 | -42.2 | n/a | n/a | n/a | n/a | n/a | 7.4 | 4 |
The Ohio State University | Assay Description The Ki values were determined by the application of the Cheng-Prusoff equation: Ki = (IC50 x Kd)/(Kd+[L]) where [L] is the concentration of [3H]MIB (... | Bioorg Med Chem Lett 18: 5567-70 (2008) Article DOI: 10.1016/j.bmcl.2008.09.002 BindingDB Entry DOI: 10.7270/Q2W0948B | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| S-ribosylhomocysteine lyase (Bacillus subtilis) | BDBM50186746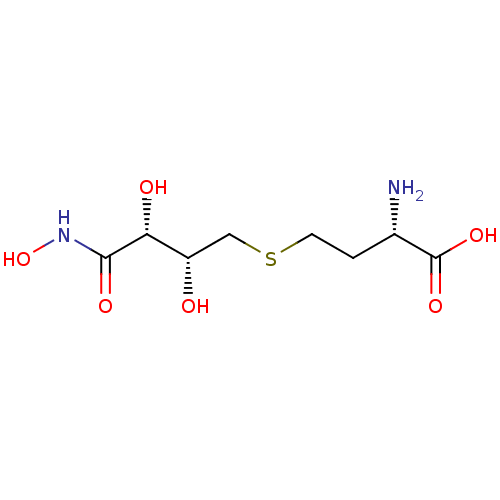 ((2S)-2-AMINO-4-[(2R,3R)-2,3-DIHYDROXY-3-N-HYDROXYC...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | 370 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibition of cobalt substituted Bacillus subtilis LuxS | J Med Chem 49: 3003-11 (2006) Article DOI: 10.1021/jm060047g BindingDB Entry DOI: 10.7270/Q25X28JQ | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| S-ribosylhomocysteine lyase (Bacillus subtilis) | BDBM50186746 ((2S)-2-AMINO-4-[(2R,3R)-2,3-DIHYDROXY-3-N-HYDROXYC...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | 430 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibition of ferrous substituted Bacillus subtilis LuxS | J Med Chem 49: 3003-11 (2006) Article DOI: 10.1021/jm060047g BindingDB Entry DOI: 10.7270/Q25X28JQ | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| S-ribosylhomocysteine lyase (Bacillus subtilis) | BDBM50186747 ((2S)-2-AMINO-4-[(2R,3S)-2,3-DIHYDROXY-3-N-HYDROXYC...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | 720 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibition of ferrous substituted Bacillus subtilis LuxS | J Med Chem 49: 3003-11 (2006) Article DOI: 10.1021/jm060047g BindingDB Entry DOI: 10.7270/Q25X28JQ | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| S-ribosylhomocysteine lyase (Bacillus subtilis) | BDBM50186747 ((2S)-2-AMINO-4-[(2R,3S)-2,3-DIHYDROXY-3-N-HYDROXYC...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | 720 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibition of cobalt substituted Bacillus subtilis LuxS | J Med Chem 49: 3003-11 (2006) Article DOI: 10.1021/jm060047g BindingDB Entry DOI: 10.7270/Q25X28JQ | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| S-ribosylhomocysteine lyase (Escherichia coli (strain K12)) | BDBM50186747 ((2S)-2-AMINO-4-[(2R,3S)-2,3-DIHYDROXY-3-N-HYDROXYC...) | PDB KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | 3.20E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibition of cobalt substituted Escherichia coli LuxS | J Med Chem 49: 3003-11 (2006) Article DOI: 10.1021/jm060047g BindingDB Entry DOI: 10.7270/Q25X28JQ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| S-ribosylhomocysteine lyase (Vibrio harveyi (strain ATCC BAA-1116 / BB120)) | BDBM50186747 ((2S)-2-AMINO-4-[(2R,3S)-2,3-DIHYDROXY-3-N-HYDROXYC...) | KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | 9.70E+3 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibition of cobalt substituted Vibrio harveyi LuxS | J Med Chem 49: 3003-11 (2006) Article DOI: 10.1021/jm060047g BindingDB Entry DOI: 10.7270/Q25X28JQ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| S-ribosylhomocysteine lyase (Bacillus subtilis) | BDBM50186746 ((2S)-2-AMINO-4-[(2R,3R)-2,3-DIHYDROXY-3-N-HYDROXYC...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | 1.06E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibition of zinc substituted Bacillus subtilis LuxS | J Med Chem 49: 3003-11 (2006) Article DOI: 10.1021/jm060047g BindingDB Entry DOI: 10.7270/Q25X28JQ | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| S-ribosylhomocysteine lyase (Escherichia coli (strain K12)) | BDBM50186746 ((2S)-2-AMINO-4-[(2R,3R)-2,3-DIHYDROXY-3-N-HYDROXYC...) | PDB KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | 1.27E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibition of cobalt substituted Escherichia coli LuxS | J Med Chem 49: 3003-11 (2006) Article DOI: 10.1021/jm060047g BindingDB Entry DOI: 10.7270/Q25X28JQ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| S-ribosylhomocysteine lyase (Vibrio harveyi (strain ATCC BAA-1116 / BB120)) | BDBM50186746 ((2S)-2-AMINO-4-[(2R,3R)-2,3-DIHYDROXY-3-N-HYDROXYC...) | KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | Article PubMed | 1.28E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibition of cobalt substituted Vibrio harveyi LuxS | J Med Chem 49: 3003-11 (2006) Article DOI: 10.1021/jm060047g BindingDB Entry DOI: 10.7270/Q25X28JQ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| S-ribosylhomocysteine lyase (Bacillus subtilis) | BDBM50186747 ((2S)-2-AMINO-4-[(2R,3S)-2,3-DIHYDROXY-3-N-HYDROXYC...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem Similars | MMDB PDB Article PubMed | 1.96E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibition of zinc substituted Bacillus subtilis LuxS | J Med Chem 49: 3003-11 (2006) Article DOI: 10.1021/jm060047g BindingDB Entry DOI: 10.7270/Q25X28JQ | |||||||||||
| More data for this Ligand-Target Pair |  3D Structure (crystal) | ||||||||||||
| S-ribosylhomocysteine lyase (Bacillus subtilis) | BDBM50142500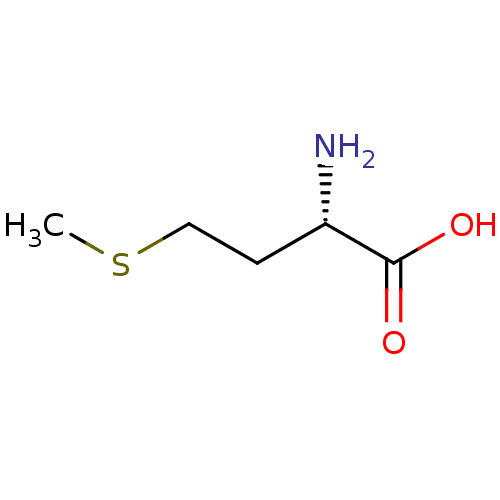 ((2S)-2-amino-4-(methylsulfanyl)butanoic acid | (S)...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | Purchase CHEMBL DrugBank MCE KEGG MMDB PC cid PC sid PDB UniChem Patents Similars | Article PubMed | 6.10E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibition of cobalt substituted Bacillus subtilis LuxS | J Med Chem 49: 3003-11 (2006) Article DOI: 10.1021/jm060047g BindingDB Entry DOI: 10.7270/Q25X28JQ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| S-ribosylhomocysteine lyase (Bacillus subtilis) | BDBM50186742 ((2S)-2-amino-6-(N-formyl-N-hydroxylamino)hexanoic ...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem | Article PubMed | 6.80E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibition of cobalt substituted Bacillus subtilis LuxS | J Med Chem 49: 3003-11 (2006) Article DOI: 10.1021/jm060047g BindingDB Entry DOI: 10.7270/Q25X28JQ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| S-ribosylhomocysteine lyase (Bacillus subtilis) | BDBM50186743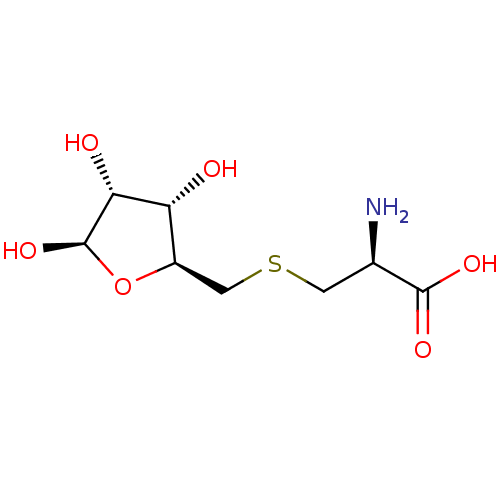 (CHEMBL383729 | S-ribosylcysteine) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL PC cid PC sid UniChem | Article PubMed | 7.00E+4 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibition of cobalt substituted Bacillus subtilis LuxS | J Med Chem 49: 3003-11 (2006) Article DOI: 10.1021/jm060047g BindingDB Entry DOI: 10.7270/Q25X28JQ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| S-ribosylhomocysteine lyase (Bacillus subtilis) | BDBM50186748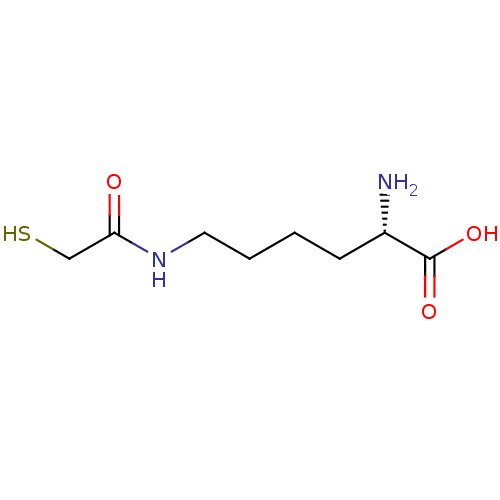 ((S)-2-amino-6-(2-mercaptoacetamido)hexanoic acid |...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL KEGG PC cid PC sid UniChem | Article PubMed | 1.32E+5 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibition of cobalt substituted Bacillus subtilis LuxS | J Med Chem 49: 3003-11 (2006) Article DOI: 10.1021/jm060047g BindingDB Entry DOI: 10.7270/Q25X28JQ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| S-ribosylhomocysteine lyase (Bacillus subtilis) | BDBM50148771 ((2R,3R)-N,2,3,4-TETRAHYDROXYBUTANAMIDE | (2R,4R)-2...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem | Article PubMed | 1.47E+5 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibition of ferrous substituted Bacillus subtilis LuxS | J Med Chem 49: 3003-11 (2006) Article DOI: 10.1021/jm060047g BindingDB Entry DOI: 10.7270/Q25X28JQ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| S-ribosylhomocysteine lyase (Bacillus subtilis) | BDBM50186744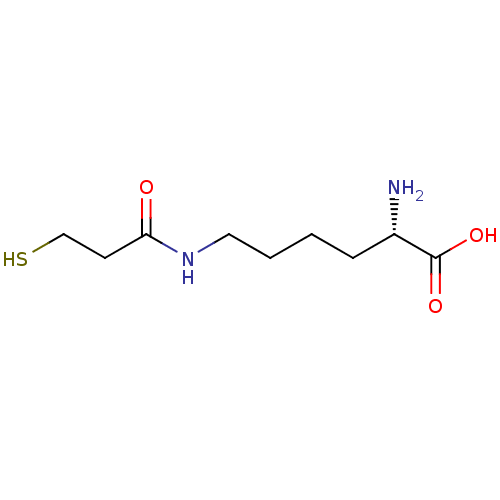 ((S)-2-amino-6-(3-mercaptopropanamido)hexanoic acid...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL KEGG PC cid PC sid UniChem | Article PubMed | 1.55E+5 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibition of cobalt substituted Bacillus subtilis LuxS | J Med Chem 49: 3003-11 (2006) Article DOI: 10.1021/jm060047g BindingDB Entry DOI: 10.7270/Q25X28JQ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| S-ribosylhomocysteine lyase (Bacillus subtilis) | BDBM50148771 ((2R,3R)-N,2,3,4-TETRAHYDROXYBUTANAMIDE | (2R,4R)-2...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem | Article PubMed | 1.56E+5 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibition of cobalt substituted Bacillus subtilis LuxS | J Med Chem 49: 3003-11 (2006) Article DOI: 10.1021/jm060047g BindingDB Entry DOI: 10.7270/Q25X28JQ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| S-ribosylhomocysteine lyase (Bacillus subtilis) | BDBM50186745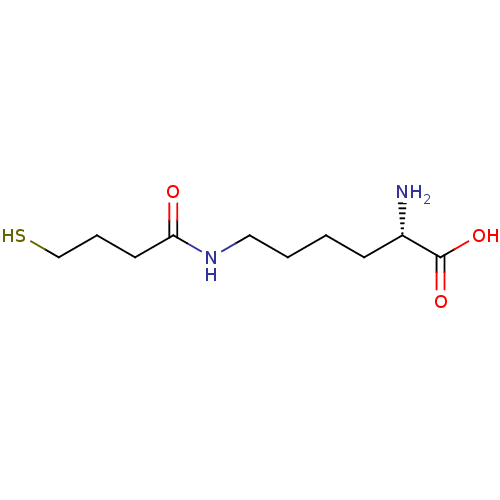 ((S)-2-amino-6-(4-mercaptobutanamido)hexanoic acid ...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL KEGG PC cid PC sid UniChem | Article PubMed | 4.73E+5 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibition of cobalt substituted Bacillus subtilis LuxS | J Med Chem 49: 3003-11 (2006) Article DOI: 10.1021/jm060047g BindingDB Entry DOI: 10.7270/Q25X28JQ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| S-ribosylhomocysteine lyase (Vibrio harveyi (strain ATCC BAA-1116 / BB120)) | BDBM50148771 ((2R,3R)-N,2,3,4-TETRAHYDROXYBUTANAMIDE | (2R,4R)-2...) | KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem | Article PubMed | 5.50E+5 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibition of cobalt substituted Vibrio harveyi LuxS | J Med Chem 49: 3003-11 (2006) Article DOI: 10.1021/jm060047g BindingDB Entry DOI: 10.7270/Q25X28JQ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| S-ribosylhomocysteine lyase (Escherichia coli (strain K12)) | BDBM50148771 ((2R,3R)-N,2,3,4-TETRAHYDROXYBUTANAMIDE | (2R,4R)-2...) | PDB KEGG UniProtKB/SwissProt GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem | Article PubMed | 7.20E+5 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibition of cobalt substituted Escherichia coli LuxS | J Med Chem 49: 3003-11 (2006) Article DOI: 10.1021/jm060047g BindingDB Entry DOI: 10.7270/Q25X28JQ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||
| S-ribosylhomocysteine lyase (Bacillus subtilis) | BDBM50148771 ((2R,3R)-N,2,3,4-TETRAHYDROXYBUTANAMIDE | (2R,4R)-2...) | PDB MMDB KEGG UniProtKB/SwissProt B.MOAD DrugBank GoogleScholar AffyNet | CHEMBL MMDB PC cid PC sid PDB UniChem | Article PubMed | 2.40E+6 | n/a | n/a | n/a | n/a | n/a | n/a | n/a | n/a |
The Ohio State University Curated by ChEMBL | Assay Description Inhibition of zinc substituted Bacillus subtilis LuxS | J Med Chem 49: 3003-11 (2006) Article DOI: 10.1021/jm060047g BindingDB Entry DOI: 10.7270/Q25X28JQ | |||||||||||
| More data for this Ligand-Target Pair | |||||||||||||